Advertisement
Research Article Free access | 10.1172/JCI73214
Disrupting SUMOylation enhances transcriptional function and ameliorates polyglutamine androgen receptor–mediated disease
Jason P. Chua,1,2,3 Satya L. Reddy,1 Zhigang Yu,1 Elisa Giorgetti,1 Heather L. Montie,4 Sarmistha Mukherjee,5 Jake Higgins,6 Richard C. McEachin,7 Diane M. Robins,6 Diane E. Merry,4 Jorge A. Iñiguez-Lluhí,5 and Andrew P. Lieberman1
1Department of Pathology,
2Neuroscience Graduate Program, and
3Medical Scientist Training Program, University of Michigan, Ann Arbor, Michigan, USA.
4Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, Pennsylvania, USA.
5Department of Pharmacology,
6Department of Human Genetics, and
7Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA.
Address correspondence to: Andrew Lieberman, Department of Pathology, University of Michigan Medical School, 3510 MSRB1, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0605, USA. Phone: 734.647.4624; E-mail: liebermn@umich.edu.
Find articles by Chua, J. in: PubMed | Google Scholar
1Department of Pathology,
2Neuroscience Graduate Program, and
3Medical Scientist Training Program, University of Michigan, Ann Arbor, Michigan, USA.
4Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, Pennsylvania, USA.
5Department of Pharmacology,
6Department of Human Genetics, and
7Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA.
Address correspondence to: Andrew Lieberman, Department of Pathology, University of Michigan Medical School, 3510 MSRB1, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0605, USA. Phone: 734.647.4624; E-mail: liebermn@umich.edu.
Find articles by Reddy, S. in: PubMed | Google Scholar
1Department of Pathology,
2Neuroscience Graduate Program, and
3Medical Scientist Training Program, University of Michigan, Ann Arbor, Michigan, USA.
4Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, Pennsylvania, USA.
5Department of Pharmacology,
6Department of Human Genetics, and
7Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA.
Address correspondence to: Andrew Lieberman, Department of Pathology, University of Michigan Medical School, 3510 MSRB1, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0605, USA. Phone: 734.647.4624; E-mail: liebermn@umich.edu.
Find articles by Yu, Z. in: PubMed | Google Scholar
1Department of Pathology,
2Neuroscience Graduate Program, and
3Medical Scientist Training Program, University of Michigan, Ann Arbor, Michigan, USA.
4Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, Pennsylvania, USA.
5Department of Pharmacology,
6Department of Human Genetics, and
7Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA.
Address correspondence to: Andrew Lieberman, Department of Pathology, University of Michigan Medical School, 3510 MSRB1, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0605, USA. Phone: 734.647.4624; E-mail: liebermn@umich.edu.
Find articles by Giorgetti, E. in: PubMed | Google Scholar
1Department of Pathology,
2Neuroscience Graduate Program, and
3Medical Scientist Training Program, University of Michigan, Ann Arbor, Michigan, USA.
4Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, Pennsylvania, USA.
5Department of Pharmacology,
6Department of Human Genetics, and
7Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA.
Address correspondence to: Andrew Lieberman, Department of Pathology, University of Michigan Medical School, 3510 MSRB1, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0605, USA. Phone: 734.647.4624; E-mail: liebermn@umich.edu.
Find articles by Montie, H. in: PubMed | Google Scholar
1Department of Pathology,
2Neuroscience Graduate Program, and
3Medical Scientist Training Program, University of Michigan, Ann Arbor, Michigan, USA.
4Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, Pennsylvania, USA.
5Department of Pharmacology,
6Department of Human Genetics, and
7Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA.
Address correspondence to: Andrew Lieberman, Department of Pathology, University of Michigan Medical School, 3510 MSRB1, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0605, USA. Phone: 734.647.4624; E-mail: liebermn@umich.edu.
Find articles by Mukherjee, S. in: PubMed | Google Scholar
1Department of Pathology,
2Neuroscience Graduate Program, and
3Medical Scientist Training Program, University of Michigan, Ann Arbor, Michigan, USA.
4Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, Pennsylvania, USA.
5Department of Pharmacology,
6Department of Human Genetics, and
7Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA.
Address correspondence to: Andrew Lieberman, Department of Pathology, University of Michigan Medical School, 3510 MSRB1, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0605, USA. Phone: 734.647.4624; E-mail: liebermn@umich.edu.
Find articles by Higgins, J. in: PubMed | Google Scholar
1Department of Pathology,
2Neuroscience Graduate Program, and
3Medical Scientist Training Program, University of Michigan, Ann Arbor, Michigan, USA.
4Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, Pennsylvania, USA.
5Department of Pharmacology,
6Department of Human Genetics, and
7Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA.
Address correspondence to: Andrew Lieberman, Department of Pathology, University of Michigan Medical School, 3510 MSRB1, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0605, USA. Phone: 734.647.4624; E-mail: liebermn@umich.edu.
Find articles by McEachin, R. in: PubMed | Google Scholar
1Department of Pathology,
2Neuroscience Graduate Program, and
3Medical Scientist Training Program, University of Michigan, Ann Arbor, Michigan, USA.
4Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, Pennsylvania, USA.
5Department of Pharmacology,
6Department of Human Genetics, and
7Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA.
Address correspondence to: Andrew Lieberman, Department of Pathology, University of Michigan Medical School, 3510 MSRB1, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0605, USA. Phone: 734.647.4624; E-mail: liebermn@umich.edu.
Find articles by Robins, D. in: PubMed | Google Scholar
1Department of Pathology,
2Neuroscience Graduate Program, and
3Medical Scientist Training Program, University of Michigan, Ann Arbor, Michigan, USA.
4Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, Pennsylvania, USA.
5Department of Pharmacology,
6Department of Human Genetics, and
7Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA.
Address correspondence to: Andrew Lieberman, Department of Pathology, University of Michigan Medical School, 3510 MSRB1, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0605, USA. Phone: 734.647.4624; E-mail: liebermn@umich.edu.
Find articles by Merry, D. in: PubMed | Google Scholar
1Department of Pathology,
2Neuroscience Graduate Program, and
3Medical Scientist Training Program, University of Michigan, Ann Arbor, Michigan, USA.
4Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, Pennsylvania, USA.
5Department of Pharmacology,
6Department of Human Genetics, and
7Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA.
Address correspondence to: Andrew Lieberman, Department of Pathology, University of Michigan Medical School, 3510 MSRB1, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0605, USA. Phone: 734.647.4624; E-mail: liebermn@umich.edu.
Find articles by Iñiguez-Lluhí, J. in: PubMed | Google Scholar
1Department of Pathology,
2Neuroscience Graduate Program, and
3Medical Scientist Training Program, University of Michigan, Ann Arbor, Michigan, USA.
4Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, Pennsylvania, USA.
5Department of Pharmacology,
6Department of Human Genetics, and
7Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA.
Address correspondence to: Andrew Lieberman, Department of Pathology, University of Michigan Medical School, 3510 MSRB1, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0605, USA. Phone: 734.647.4624; E-mail: liebermn@umich.edu.
Find articles by Lieberman, A. in: PubMed | Google Scholar
Published January 20, 2015 - More info
J Clin Invest. 2015;125(2):831–845. https://doi.org/10.1172/JCI73214.
Copyright © 2015, American Society for Clinical Investigation
Received: September 13, 2013; Accepted: November 25, 2014
Related article:
Abstract
Spinobulbar muscular atrophy (SBMA) is an X-linked disease characterized by degeneration of motor neurons, muscle atrophy, and progressive weakness. It is caused by a polyglutamine (polyQ) expansion in the androgen receptor (AR), a transcription factor that is activated upon hormone binding. The polyQ expansion in AR causes it to form intracellular aggregates and impairs transcriptional activity. Intriguingly, SUMOylation (where SUMO indicates small ubiquitin-like modifier) of AR inhibits its transcriptional activity and reduces aggregation of the polyQ form of this protein, but it is unclear whether SUMOylation plays a pathogenic or protective role in SBMA. In this issue of the
Authors
Tim J. Craig, Jeremy M. Henley
-
Abstract
Expansion of the polyglutamine (polyQ) tract within the androgen receptor (AR) causes neuromuscular degeneration in individuals with spinobulbar muscular atrophy (SBMA). PolyQ AR has diminished transcriptional function and exhibits ligand-dependent proteotoxicity, features that have both been implicated in SBMA; however, the extent to which altered AR transcriptional function contributes to pathogenesis remains controversial. Here, we sought to dissociate effects of diminished AR function from polyQ-mediated proteotoxicity by enhancing the transcriptional activity of polyQ AR. To accomplish this, we bypassed the inhibitory effect of AR SUMOylation (where SUMO indicates small ubiquitin-like modifier) by mutating conserved lysines in the polyQ AR that are sites of SUMOylation. We determined that replacement of these residues by arginine enhances polyQ AR activity as a hormone-dependent transcriptional regulator. In a murine model, disruption of polyQ AR SUMOylation rescued exercise endurance and type I muscle fiber atrophy; it also prolonged survival. These changes occurred without overt alterations in polyQ AR expression or aggregation, revealing the favorable trophic support exerted by the ligand-activated receptor. Our findings demonstrate beneficial effects of enhancing the transcriptional function of the ligand-activated polyQ AR and indicate that the SUMOylation pathway may be a potential target for therapeutic intervention in SBMA.
-
Introduction
The CAG/polyglutamine (CAG/polyQ) disorders are a family of 9 neurodegenerative diseases caused by similar microsatellite expansions in coding regions of unrelated genes (1). Among these diseases is spinobulbar muscular atrophy (SBMA), a progressive neuromuscular disorder that occurs only in men and is characterized by proximal limb and bulbar muscle weakness, atrophy, and fasciculations (2). Clinical onset of SBMA occurs in adolescence to adulthood and is characterized initially by muscle cramps and elevated serum creatine kinase (3, 4). These myopathic features commonly precede muscle weakness, which inevitably develops as the disease progresses. The causative mutation in SBMA is an expansion of a CAG repeat in the first exon of the androgen receptor (AR) gene (5). The expanded glutamine tract promotes hormone-dependent AR unfolding and oligomerization, steps that are critical to toxicity. In SBMA, as in other CAG/polyQ disorders, the mutant protein disrupts multiple downstream pathways and toxicity likely results from the cumulative effects of altering a diverse array of cellular processes including transcription, RNA splicing, axonal transport, and mitochondrial function (6–15). Mechanisms that lead to the selective impairment of the neuromuscular system remain incompletely understood, yet recent studies have established that peripheral expression of the polyQ AR in skeletal muscle is an important contributor to this process (16, 17).
In addition to the proteotoxic gain of function, polyQ expansion also negatively affects the intrinsic transcriptional activity of the AR (6, 8–10). While SBMA patients exhibit signs of partial androgen insensitivity, as manifested by diminished fertility and gynecomastia, the extent to which loss of AR function contributes to pathogenesis remains poorly understood, mainly because of the difficulty in dissociating effects of proteotoxicity from impaired intrinsic transcriptional function. To explore this question, we sought to define the consequences of mitigating the transcriptional deficits of the polyQ AR by relieving the inhibitory effect of AR posttranslational modification by small ubiquitin-like modifier (SUMO) (18, 19).
SUMO proteins share with ubiquitin a common structural fold and are reversibly conjugated to lysine residues in target proteins through an enzymatic pathway analogous to ubiquitination, but carried out by a distinct set of enzymes (20). AR was one of the first proteins shown to be subject to SUMOylation (19), and this modification occurs at 2 lysines (K385, K518) in the N-terminal region. These residues are embedded within short amino acid motifs that have been functionally defined in AR and other sequence-specific factors as exerting a promoter context inhibitory effect (18). As is the case for most proteins, steady-state stoichiometry of SUMOylation of AR is low, likely because of active reversal by SUMO proteases such as SUMO1/sentrin-specific peptidase 1 (SENP1) (21). Nevertheless, transient SUMOylation exerts strong inhibitory effects through a distinct surface in SUMO (22) that is recognized by SUMO-binding motifs in partner proteins (23). Although the exact nature of the SUMO-binding factors responsible for the inhibitory effects on AR is not fully defined, mechanistic studies indicate that SUMOylation exerts its effects preferentially on stably bound receptor-DNA complexes (24) and involves a redistribution of AR away from chromatin (25).
SUMOylation has attracted considerable attention for its impact on neurodegenerative diseases, including polyQ disorders (26, 27). In addition to AR, several mutant proteins that cause neurodegeneration are targets of SUMOylation. The extent to which SUMO influences disease pathogenesis varies in a context-dependent manner (28). Here, we take advantage of the modulatory role of SUMOylation on AR function to test the hypothesis that diminished transcriptional activity of the polyQ AR contributes to SBMA pathogenesis. In this view, restoring transcriptional function would be expected to attenuate some aspects of the disease. Our approach is based on the established inhibitory effects of SUMO on AR function, the significant loss of transactivation caused by polyQ expansion (6–10, 29), and prior studies suggesting that native AR function contributes to SBMA pathogenesis (30). To test our hypothesis, we used gene targeting to generate a knockin mouse line that expresses a polyQ-expanded AR that cannot be SUMOylated. Our studies characterizing this new mouse model of SBMA, complemented by additional experimental systems in vitro, provide a unique avenue for examining the effects of restoring polyQ AR function and serve to dissociate the pathogenic role of AR dysfunction from polyQ proteotoxicity in vivo.
-
Results
Establishment of a cellular model of a SUMOylation-resistant polyQ AR. To study the effects of abrogating SUMOylation of the polyQ AR, we initially generated PC12 cells expressing tetracycline-regulated AR10Q, AR112Q, and AR111Q, with its 2 SUMO-acceptor lysine residues mutated to arginine (AR111Q-KRKR) (Figure 1A). With these cells, we tested to determine whether introducing mutations that prevent SUMOylation affected the response of polyQ AR to ligand. In the absence of the synthetic nonaromatizable androgen R1881, all 3 variants of AR displayed a predominantly cytoplasmic distribution, in agreement with expected localization when unbound to ligand (Figure 1, B and C). Upon addition of R1881, all 3 AR forms accumulated in the nucleus with comparable kinetics (Figure 1B) and both AR112Q and AR111Q-KRKR formed ligand-dependent intranuclear aggregates (Figure 1C). To verify that the KRKR mutations interrupt AR SUMOylation, we transiently transfected AR112Q and AR111Q-KRKR cells with HA-tagged SUMO3, then immunoprecipitated each polyQ AR variant and probed for HA. Western blot analysis confirmed that polyQ AR remained a target of SUMOylation and that the KRKR mutation prevented this modification (Figure 1D), in line with previous transient expression studies (31). In contrast, the KRKR mutation did not alter ubiquitination (Figure 1E), another lysine-directed posttranslational modification of the polyQ AR. Both AR112Q and AR111Q-KRKR were ubiquitinated in cells expressing the human E3 ligase C terminus of Hsc70-interacting protein (hCHIP) and HA-tagged ubiquitin. Moreover, ubiquitination of both AR variants was enhanced following treatment with YM-1 (Figure 1E), an allosteric regulator of Hsp70 that increases polyQ AR degradation (32). These data indicate that the KRKR mutation abolishes SUMO modification of the polyQ AR without significantly altering ligand-dependent localization or ubiquitination.
 Figure 1
Figure 1A cellular model expressing non-SUMOylatable polyQ AR. (A) AR10Q, AR112Q, or AR111Q-KRKR were induced to express AR in the presence of 10 nM R1881 for 24 hours, then analyzed by Western blot. Dox, doxycycline. (B and C) AR10Q, AR112Q, and AR111Q-KRKR cells were induced to express AR for 24 hours, then treated with R1881 for varying times and immunostained for AR (red). Nuclei were stained by DAPI. (B) Cytoplasmic (score 0) to nuclear (score 4) translocation was determined at indicated times. Data are mean ± SEM. n = 3 replicate experiments. (C) Arrows indicate nuclei containing aggregates after 24 hours of treatment with 10 nM R1881. Scale bar: 2 μm. (D) AR112Q- and AR111Q-KRKR–expressing cells were transfected with HA-SUMO3 and treated with 10 nM R1881 for 24 hours. Whole-cell lysates (left panel) were immunoprecipitated for AR and blotted for HA or AR (right panel). (E) AR112Q or AR111Q-KRKR cells were transfected with HA-ubiquitin and hCHIP, and treated with YM-1, as indicated. Following incubation with 10 mM MG132, lysates were collected and analyzed for AR ubiquitination. Whole-cell lysates (left panel) were immunoprecipitated for AR and blotted for AR and HA (middle and right panels).
Preventing SUMOylation enhances transcriptional activity of the polyQ AR. To examine the effects of the KRKR mutation on polyQ AR transcriptional function, PC12 cells were induced to express similar levels of AR10Q, AR112Q, or AR111Q-KRKR (as in Figure 1A) and then stimulated with R1881 or vehicle control for 24 hours. Triplicate RNA samples from each line were collected, and the corresponding cDNAs were analyzed by Affymetrix GeneChip microarrays (Figure 2, A–C). A heat map of the 93 genes significantly activated (in red) or repressed (in blue) by androgen in AR10Q cells and the corresponding levels in AR112Q and AR111Q-KRKR is shown in Figure 2A. This analysis confirmed a glutamine length–dependent partial loss of AR function with a significant reduction in the amplitude of regulation for most genes (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI73214DS1). Of the 93 ligand-responsive genes identified in cells expressing AR10Q, only 32 met criteria for ligand regulation (≥1.5-fold change) in AR112Q cells (Figure 2C). Detailed analyses revealed that the regulatory deficits in AR112Q cells were broad across the set of 93 ligand-responsive genes and affected genes irrespective of the relative abundance of their transcripts (not shown). Notably, and consistent with the promoter context-dependent effects of AR SUMOylation, disruption of the SUMOylation sites partially counteracted this polyQ-induced loss of function. AR111Q-KRKR cells showed enhanced responsiveness to a subset of genes, increasing the number of genes meeting the threshold for differential expression to 49 (Figure 2C). To confirm these data, we analyzed selected genes by quantitative real-time RT-PCR (qPCR). The Ppard gene, drawn from the microarray subset of genes regulated in all 3 lines, displayed comparable induction when analyzed by qPCR (Figure 2D). In contrast, examination of 2 independent genes from the subset regulated only by AR10Q and AR111Q-KRKR (Baiap2, Chst1) confirmed a glutamine length–dependent loss of inducibility that was reversed and even further enhanced by disruption of the SUMOylation sites (Figure 2D). This same pattern of androgen-responsive changes was observed in PC12 cells expressing AR10Q and AR10Q-KRKR (Figure 2E). AR10Q and AR10Q-KRKR displayed similar ligand-dependent activation of Ppard, whereas AR10Q-KRKR triggered enhanced activation of Baiap2 and Chst1. These data are consistent with prior studies using reporter gene assays, where disrupting polyQ AR SUMOylation by introduction of the KRKR mutations or I384N/V517N substitutions or by deletion of the SUMOylation motifs enhanced activation of AR- responsive reporter gene constructs (31).
 Figure 2
Figure 2Disruption of polyQ AR SUMOylation potentiates AR function. (A) Heat map showing expression of genes activated (red) or repressed (blue) 1.5-fold or more by AR10Q cells compared with expression in AR112Q and AR111Q-KRKR cells. (B) Heat map showing expression of 183 androgen-responsive genes in AR111Q-KRKR cells that change less than 1.5-fold in AR10Q and AR112Q cells. (C) Venn diagram illustrating the number and overlap of androgen-responsive genes in AR10Q (black circle), AR112Q (blue circle), and AR111Q-KRKR cells (red circle). (D) Expression of candidate genes (Ppard, Baiap2, and Chst1 in black, red, and purple, respectively) identified in A was analyzed by qPCR. Data shown are mean ± SEM (n = 3). ****P < 0.0001 by ANOVA. (E) Expression of candidate genes (Ppard, Baiap2, and Chst1 in black, red, and purple, respectively) in AR10Q and AR10Q-KRKR cells was analyzed by qPCR. Data shown are mean ± SEM (n = 3). **P < 0.01; ***P < 0.001; ****P < 0.0001 by ANOVA.
Notably, we also found that the KRKR mutation enhanced the total number of genes meeting threshold for ligand responsiveness to 246 (Figure 2C). For the 183 genes that met the threshold for only AR111Q-KRKR, a comparison with the corresponding levels in AR10Q and AR112Q (Figure 2B) revealed that a large majority of these genes displayed ligand regulation but did not change by 1.5-fold or more (Supplemental Table 2). This outcome is consistent with the predicted enhancement in responsiveness to a subset of genes natively regulated by AR as a consequence of relief from the inhibitory effects of SUMO. These findings are also similar to those recently reported in a prostate cancer cell line where the KRKR mutation (in non–polyQ-expanded AR) was found to expand the number of induced and repressed genes (33). Taken together, these results demonstrate that, although expansion of the glutamine tract diminishes the responsiveness of genes normally regulated by AR, preventing SUMOylation through the KRKR mutation partially counteracts these deleterious effects.
Generation of AR113Q-KRKR knockin mice. To investigate the influence of SUMOylation in vivo, we used gene targeting in order to generate knockin mice that express the polyQ AR, with K385 and K518 both mutated to arginines. We modified the targeting vector used previously to generate our existing AR113Q line of knockin mice by introducing the KRKR mutations (Figure 3A). We verified correct recombination of the targeted exon 1 into the mouse AR gene by Southern blot analysis in JM8.F6 cells, an embryonic stem (ES) cell line from the C57BL/6N strain (Figure 3B). After germline transmission of the targeted allele from chimeric founders to F1 heterozygous females, we crossed these females to C57BL/6J WT males and assessed subsequent inheritance of the AR113Q-KRKR allele by PCR (Figure 3C). For the purposes of phenotypic characterization in this study, we used AR113Q male mice backcrossed for more than 10 generations to the C57BL/6J background, N1 AR113Q-KRKR male mice, and WT littermate males from both lines. Females from both AR113Q lines were similarly fertile, and both AR113Q alleles were inherited in the expected Mendelian ratios (data not shown). Male mice from the AR113Q and AR113Q-KRKR lines were indistinguishable prior to disease onset at sexual maturity.
 Figure 3
Figure 3Generation of AR113Q-KRKR knockin mice. (A) Schematic representation depicting AR113Q-KRKR targeting vector. Sphl restriction sites, 5′ and 3′ probes for Southern blot analysis, glutamine tract, lysine-to-arginine mutations, and floxed neomycin resistance cassette are indicated. Diagram modified from ref. 55. (B) Southern blot analysis of ES cell DNA after Sphl digestion with the 5′ and 3′ probes indicated in A before (lanes 1 and 4) and after (lanes 2 and 5) homologous recombination. Lanes 3 and 6 contain control liver DNA from previously targeted AR113Q knockin line. L, ladder. (C) PCR primers were used to amplify sequences from a WT female (lane 1), F1 AR113Q-KRKR female (lane 2), and N1 AR113Q-KRKR male (lane 3) produced by the female in lane 2.
Preventing AR SUMOylation alters ligand-dependent gene regulation in vivo. We initially sought to determine the consequences of disrupting AR SUMOylation on ligand-dependent gene regulation in vivo. To accomplish this, a cohort of WT, AR113Q, and AR113Q-KRKR adult males were surgically castrated in order to suppress endogenous ligand activation of AR. Three weeks after orchiectomy, animals were treated with testosterone propionate or vehicle by i.p. injection and tissues were harvested after 16 hours. The expression of well-characterized androgen-induced genes in prostate was examined by qPCR (Figure 4A). Our analysis showed that the strong ligand-induced expression of Nkx3-1 in WT males was absent in the AR113Q line, but was fully restored in AR113Q-KRKR mice. A similar pattern was also observed for Fkbp5. In the case of Pbsn, the modest and not significant induction in WT males was absent in ARQ113 mice, but was strongly enhanced in AR113Q-KRKR mice. To determine effects on androgen-regulated gene expression in skeletal muscle, we harvested the levator ani-bulbocavernosus (LABC), a muscle that expresses high levels of AR. We used an AR-signaling target array to identify candidate androgen-responsive genes in muscle from WT mice (data not shown). We selected 7 genes that were repressed by ligand treatment and confirmed that all transcripts were significantly decreased in muscle following androgen treatment of WT males (Figure 4B). In AR113Q males, none of these genes responded to androgen treatment, whereas appropriate regulation was observed in AR113Q-KRKR males. Taken together, these data are consistent with our PC12 cellular model and demonstrate that disrupting polyQ AR SUMOylation restores acute androgen responsiveness to AR target genes in vivo.
 Figure 4
Figure 4Expression of androgen-responsive genes in prostate and skeletal muscle. WT (black bars), AR113Q (white bars), and AR113Q-KRKR (gray bars) adult males underwent orchiectomy and then 3 weeks later were treated with 200 mg testosterone propionate i.p. (+) or vehicle (–). Prostate (A) and LABC muscle (B) were harvested after 16 hours. Expression of AR-responsive genes was analyzed by qPCR. Data shown are mean ± SEM (n = 3/group). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant by ANOVA.
AR113Q-KRKR males demonstrate partial androgen compromise. Since partial loss of AR function and features of androgen insensitivity are molecular and phenotypic hallmarks of SBMA, we examined several measures of androgen function on the hypothalamic-pituitary-gonadal (HPG) axis in AR113Q-KRKR mice at 13 to 15 weeks of age. Compared with WT mice, both AR113Q and AR113Q-KRKR mice demonstrated testicular and seminal vesicle atrophy and reduced sperm counts (Figure 5A). Modest elevations in serum testosterone and luteinizing hormone (LH) were evident in both AR113Q and AR113Q-KRKR males, but because of variability between animals, these changes did not reach statistical significance (Figure 5B). We also analyzed testicular expression of the gene encoding 3-β-hydroxysteroid dehydrogenase type I (3βHSD-I, Hsd3b1), an enzyme responsible for ketosteroid hormone synthesis in Leydig cells in response to LH and whose expression is indicative of androgen insensitivity (34). Consistent with the endocrine tissue pathology and serum studies, we found similar increases in Hsd3b1 mRNA in testes from both 113Q lines of mice (Figure 5C). These data demonstrate that compromise of the androgen axis remains evident when polyQ AR SUMOylation is prevented. Furthermore, we speculated that chronically enhanced AR activity in the KRKR mutant could lead to inhibition of the HPG axis in aged males. Consistent with this notion, adult AR113Q-KRKR males at 7 to 9 months exhibited more severe testicular atrophy and decreased sperm counts compared with similarly aged AR113Q males (Figure 5D).
 Figure 5
Figure 5Characterization of the HPG axis in AR113Q-KRKR mice. (A) Testes and seminal vesicles were collected at 13 to 15 weeks and analyzed by weight (left and middle panels). Epididymes were collected and sperm count determined (right panel). Data are shown as mean ± SEM (n = 6 per group). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by ANOVA. (B) Serum samples (n = 6 per group) were collected at 13 to 15 weeks and analyzed for levels of testosterone (T) and LH. Data are shown as mean ± SEM. (C) Total RNA was extracted from testes from WT, AR113Q, and AR113Q-KRKR mice (n = 3 per group) at 13 to 15 weeks and analyzed for Hsd3b1 expression by qPCR. Data are shown as mean ± SEM. ****P < 0.0001 by ANOVA. (D) Testis weight and epididymal sperm count from males at 7 to 9 months. Data are shown as mean ± SEM. *P < 0.05; ***P < 0.001; ****P < 0.0001 by ANOVA.
Behavioral and neuromuscular phenotypes in AR113Q-KRKR mice. As the phenotype of AR113Q mice includes decreased body mass and grip strength, we also evaluated these parameters in AR113Q-KRKR mice and found indistinguishable deficits (Figure 6, A and B). The reduction in these measures became evident at approximately 12 weeks and persisted thereafter. To broaden our characterization of muscle function beyond assessment of grip strength, we examined exercise capacity by treadmill running until exhaustion. By this measure we detected a significant decrement in exercise endurance in AR113Q mice (Figure 6C), consistent with other measures of functional decline in SBMA muscle (35, 36). Strikingly, KRKR mutations in the polyQ AR yielded a profound rescue of exercise capacity to levels equivalent to those of WT mice. This functional rescue persisted in mice aged up to 9 months (Figure 6D). Since many trophic effects of androgens on muscle are thought to be a consequence of ligand-activated gene regulation, we hypothesized that the recovery in KRKR mutants reflected a restored trophic support that would be dependent upon ligand. To test this possibility, we applied the same exercise paradigm to AR113Q-KRKR males that had been castrated at 5 to 6 weeks and to WT females. Exercise trials of these 2 cohorts revealed substantially less exercise capacity compared with gonadally intact males regardless of glutamine tract length (Figure 6C). These results confirmed the supportive role of androgens in the development of exercise endurance and in directing the rescue in SUMO-resistant SBMA mice.
 Figure 6
Figure 6Phenotypic characterization of AR113Q-KRKR knockin mice. (A) WT, AR113Q, and AR113Q-KRKR male mice (n = 10 per group) were weighed and (B) assessed for forelimb grip strength every 2 weeks. Data shown are mean ± SEM. (C) WT (n = 7), AR113Q (n = 6), AR113Q-KRKR male mice (n = 6), castrated AR113Q-KRKR males (AR113Q-KRKR-C, n = 4), and WT females (n = 4) were exercised until exhaustion at 13 to 15 weeks of age, and total time and distance run were recorded. Data are mean ± SEM. *P < 0.05; **P < 0.01 by ANOVA. (D) WT, 113Q, and 113Q-KRKR (n = 3 per group) males were exercised until exhaustion at 7 to 9 months of age, and total time and distance run were recorded. Data are mean ± SEM. *P < 0.05 by ANOVA. (E) WT, AR113Q, and AR113Q-KRKR male mice (n = 10 per group) were followed for overall survival (left panel, P < 0.0001 113Q vs. 113Q-KRKR by Mantel-Cox log-rank test), and both 113Q knockin mouse lines were monitored for disease onset (middle panel, P = 0.6) and disease duration (right panel, P < 0.02).
Since early death is another phenotypic feature of AR113Q mice, we followed cohorts of WT, AR113Q, and AR113Q-KRKR mice and monitored survival rates. In line with previous characterization (36), AR113Q mice demonstrated a precipitous decline in survival beginning at 13 weeks (Figure 6E). None of these mice survived past 40 weeks (n = 10). In contrast, AR113Q-KRKR mice exhibited a profound extension of life span. Through 40 weeks, only 1 death occurred in the experimental cohort (n = 10). This rescue of early death was significant (P < 0.0001) and took place despite a lack of discernible differences in disease onset between the SUMOylation-competent and SUMOylation-deficient AR113Q lines. We defined disease onset as the age at which grip strength of mutant males was 10% less than the mean of age-matched WT controls and found no difference between AR113Q and AR113Q-KRKR males (Figure 6E). Accordingly, the equivalent disease onset coupled with life span extension resulted in a significant extension of disease duration (P < 0.02) in the SUMOylation-deficient SBMA mice (Figure 6E). Taken together, these results show that abrogating polyQ AR SUMOylation, which enhances AR transcriptional function, exerts profound and long-lasting beneficial effects on exercise endurance and survival.
Rescue of type I fiber atrophy and gene-expression changes in AR113Q-KRKR muscle. To explore the mechanism underlying phenotypic rescue in AR113Q-KRKR mice, we initially sought to determine whether mutant forms of the AR were equivalently expressed. Immunoprecipitation (IP) of AR from quadriceps muscle and spinal cord demonstrated that levels of AR113Q and AR113Q-KRKR were similar (Figure 7A and Supplemental Figure 1). Comparable levels of AR protein were also detected in testis of WT, AR113Q, and AR113Q-KRKR mice (Figure 7B and Supplemental Figure 1). Since AR113Q mice display frequent accumulation of intranuclear aggregates of polyQ AR in skeletal muscle (36), we analyzed sections of LABC and quadriceps muscle from mice aged 13 to 15 weeks. While quadriceps nuclei in WT muscle rarely showed punctate AR intranuclear staining (5.5% ± 2.3%), nuclei from both AR113Q and AR113Q-KRKR quadriceps muscle showed substantially higher and similar amounts of polyQ AR aggregates (26.9% ± 2.2% and 29.4% ± 1.9%, respectively) (Figure 7C). A similar pattern was observed in the LABC. These analyses indicate that phenotypic rescue was not attributable to diminished expression or aggregation of AR113Q-KRKR.
 Figure 7
Figure 7Expression and aggregation of AR113Q and AR113Q-KRKR are similar. (A) AR expression in quadriceps and spinal cord from WT, AR113Q, and AR113Q-KRKR mice was analyzed by IP and Western blot. GAPDH shows similar input protein levels. (B) AR expression in testis. Hsp90 controls for loading. (C) Immunofluorescence staining for AR (red) in quadriceps and LABC. Nuclei were stained by DAPI (blue). Scale bar: 25 μm. At right, percentage of nuclei with AR-reactive puncta in the quadriceps. Data are mean ± SEM. n = 3 mice per genotype. ****P < 0.0001 by ANOVA.
Rescue of exercise endurance and survival was also not associated with attenuation of muscle atrophy or skeletal muscle gene-expression changes indicative of denervation. The tibialis anterior (TA) muscle showed significant atrophy in both AR113Q and AR113Q-KRKR mice compared with WT (Figure 8A). Both AR113Q lines expressed equivalent levels of AR mRNA and showed increased expression of acetylcholine receptor-α subunit and myogenin mRNAs in quadriceps muscle. A similar trend was observed for the expression of Myod mRNA (Figure 8B). Unexpectedly, these measurements of muscle pathology were slightly accentuated in AR113Q-KRKR mice despite rescue of exercise capacity and survival. This suggested that alterations in other muscle properties mediated the beneficial response to ligand activation of AR113Q-KRKR.
 Figure 8
Figure 8Rescue of type I fiber atrophy in AR113Q-KRKR males. (A) TA muscles were harvested from both hind limbs and weighed (n = 6 per group). Data are mean ± SEM. **P < 0.01; ****P < 0.0001 by ANOVA. (B) AR (Ar), nicotinic acetylcholine receptor-a subunit (Chrna1), myogenin (Myog), and MyoD (Myod) mRNA were analyzed in quadriceps by qPCR (n = 6 per group). *P < 0.05; ***P < 0.001; ****P < 0.0001 by ANOVA. (C) Representative type I slow-twitch heavy-chain myosin (green) and type II fast-twitch heavy-chain myosin (red) staining in soleus. Basement membrane stained with wheat germ agglutinin (green). Scale bar: 50 μm. (D) Mean area ± SEM and fiber size distribution of type I and type II fibers (n = 3 mice per group, ≥75 fibers per mouse). **P < 0.01; ****P < 0.0001 by ANOVA.
An important determinant of exercise performance and metabolic profile is the fiber type composition of skeletal muscle. Type I (slow twitch) fibers are rich in mitochondria, fatigue resistant, and reliant on oxidative metabolism, while type II (fast twitch) fibers are mitochondria poor, fatigue susceptible, and dependent on glycolytic metabolism (37, 38). Given the notable androgen-dependent improvement in exercise endurance in AR113Q-KRKR males, we performed immunostaining for fiber type–specific heavy-chain myosins on soleus muscle because of its mixed fiber type composition. In contrast, both the TA and quadriceps muscles are type II fiber predominant (39, 40). In AR113Q soleus muscle, we observed atrophy across all fiber types (Figure 8, C and D). In contrast, soleus from AR113-KRKR mice contained type I fibers that were remarkably spared from atrophy (Figure 8, C and D). Quantification of cross-sectional area demonstrated a significant rescue in the distribution and mean type I fiber size (Figure 8D). This rescue of muscle atrophy was not seen in type II fibers of AR113Q-KRKR muscle, nor was it observed in type I fibers of AR113Q-KRKR mice following castration (Figure 8D). These data reveal that the improved exercise endurance in AR113Q-KRKR was associated with rescue of type I fiber atrophy and suggest that this fiber-selective effect is mediated by trophic support from the KRKR receptor.
To explore whether AR113Q-KRKR also affects muscle predominantly composed of type II fibers despite the absence of morphological rescue of atrophy, we used RNA-Seq to analyze gene expression in quadriceps muscle from WT, AR113Q, and AR113Q-KRKR males at 14 weeks. Pairwise comparisons enabled the identification of differentially expressed genes in WT versus AR113Q muscle and WT versus AR113Q-KRKR muscle. We queried these lists against a publicly available database of literature-derived androgen-responsive genes that includes 993 mouse genes (41). This strategy identified 107 differentially expressed androgen-response genes in quadriceps muscle of WT versus AR113Q males (Figure 9A and Supplemental Table 3). Ninety-six of these genes were also differentially expressed in WT versus AR113Q-KRKR muscle. As shown in the heat map (Figure 9B), similar expression changes were identified for the majority of these genes in AR113Q and AR113Q-KRKR muscle, though AR113Q-KRKR muscle generally showed a greater magnitude of change. Interestingly, this analysis also identified an additional 221 genes that were differentially expressed in AR113Q-KRKR, but not AR113Q muscle (Figure 9, A and B), suggesting that the mutation had expanded the set of AR-regulated genes in skeletal muscle much as it had done in vitro (Figure 2). Pathway analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) revealed that these 221 genes were highly enriched for mitochondria-related functional categories (Table 1), indicating that altered expression of genes regulating energy metabolism correlated with enhanced exercise endurance. Taken together, these data suggest that disruption of AR113Q SUMOylation promotes hormone-dependent trophic effects on skeletal muscle mediated by enhanced activity of the receptor as a transcriptional regulator.
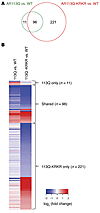 Figure 9
Figure 9Gene-expression differences in quadriceps muscle of WT, AR113Q, and AR113-KRKR males. Muscle was harvested from 14-week-old males (n = 3/genotype), and gene expression was analyzed by RNA-Seq. (A) Venn diagram illustrating the number and overlap of differentially expressed (≥ 1.5-fold) androgen-responsive genes in WT versus AR113Q (green circle) and WT versus AR113Q-KRKR (red circle). (B) Heat map of differentially expressed androgen-responsive genes in WT versus AR113Q or AR113Q-KRKR muscle.
 Table 1
Table 1Significantly enriched Gene Ontology categories related to mitochondria and energy metabolism
-
Discussion
In this study of SBMA pathogenesis, we sought to disentangle polyQ length–dependent proteotoxicity from the partial loss of AR function to discern the relative contribution of each mechanism to the disease. To accomplish this goal, we selectively counteracted functional alterations in the polyQ AR by relieving the transcriptional inhibitory effect of SUMOylation. We show that mutation of the SUMO-acceptor sites in AR exon 1 specifically prevents SUMOylation of polyQ AR without altering ligand-induced nuclear translocation, formation of intranuclear inclusions, or steady-state protein levels. Consistent with the established repressive role of SUMO on AR function, the SUMOylation-deficient polyQ AR displays broadly enhanced transcriptional activity in both cell culture and mice, with a profile that partially counteracts the polyQ-mediated loss of function. Male mice generated by gene targeting to express AR113Q-KRKR develop normally and express a phenotype only after reaching sexual maturity.
Intriguingly, AR113Q-KRKR male mice exhibit a substantial recovery of exercise capacity compared with AR113Q males. This rescue persists as animals age despite decrements in exercise fitness across all genotypes. Moreover, this rescue is hormone dependent and is associated with recovery of type I muscle fiber size, demonstrating that ligand-dependent activation of AR113Q-KRKR provides marked trophic support to muscle. It is likely that ligand activation of AR113Q-KRKR exerts beneficial effects on type II muscle fibers as well, despite the lack of rescue of fiber atrophy. This conclusion is supported by the identification of numerous gene-expression differences in quadriceps muscle of AR113Q-KRKR versus WT males that are not similarly detected in AR113Q muscle, many of which cluster in mitochondria-related pathways. Included in the list of genes selectively upregulated in AR113Q-KRKR muscle are those encoding the mitochondrial proteins translocase of mitochondrial membrane 20 homolog (TOMM20), NADH dehydrogenase (ubiquinone) Fe-S protein 3 (NDUFS3), hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase, β subunit (HADHB), cytochrome c, and cytochrome oxidase subunit VIIc. The beneficial effects of the KRKR mutation within skeletal muscle are consistent with recent studies demonstrating a critical role of peripheral polyQ AR expression in SBMA pathogenesis (16). The KRKR mutation within the polyQ AR also leads to an almost complete rescue of early death and a marked extension of disease duration. As the phenotypic rescue of AR113Q-KRKR males occurs without evidence of diminished polyQ AR expression or aggregation and is hormone dependent, we conclude that the beneficial effects of abrogating SUMOylation are mediated by enhanced transcriptional function of the mutant receptor. Notably, the conclusion that ligand activation of the receptor’s transcriptional function is beneficial to SBMA mice is distinct from that drawn from an analysis of a Drosophila model of SBMA in which native function of the AR was implicated as indispensable for disease pathogenesis (30). In contrast, our analyses demonstrate that disrupting polyQ AR SUMOylation enhances its transcriptional function and rescues key aspects of SBMA pathology. These results argue against a purely propathogenic role of native AR function in disease. As Drosophila lack endogenous androgen-responsive genes, this model is likely insensitive to trophic actions mediated by the ligand-activated receptor.
Not all aspects of the phenotype of AR113Q males are rescued in AR113Q-KRKR mutants. Disruption of SUMOylation fails to significantly modify certain aspects of muscle pathology, including weight of muscles that are primarily composed of type II fibers, grip strength, and gene-expression changes indicative of denervation. Although type I fibers are only a small fraction of overall skeletal muscle mass in mice, they are much more abundant in humans (42), suggesting that strategies to target AR SUMOylation may have greater beneficial effects in SBMA patients. It is also likely that trophic effects of the ligand-activated AR113Q-KRKR are beneficial to additional cell types and that these actions could contribute to overall health of SBMA mice. This suggests that activating androgen-regulated trophic pathways that are stimulated by the KRKR receptor may be beneficial to SBMA patients. A potential approach would be to suppress SUMO modification of AR by inhibiting the activity of E3 ligases, such as members of the protein inhibitor of activated STAT (PIAS) family (43), or by stimulating the activity of SENP1, the main SUMO peptidase responsible for deconjugating SUMO from AR (44). We are actively pursuing the discovery of agents that can regulate the activity of these enzymes. Alternatively, selective AR modulators (SARMS) that exert strong anabolic effects in skeletal muscle, but show limited activity in other organs, could be considered. Whether such SARMS can provide beneficial effects to muscle in a manner that outweighs ligand-dependent proteotoxicity awaits future testing.
Unexpectedly, endocrine pathology, including seminal vesicle and testicular atrophy, sperm count, and Hsd3b1 mRNA expression are unaffected by AR SUMOylation. These findings raise the possibility that proteotoxicity of the polyQ AR, and not simply partial loss of transactivation function, contributes to deficits in the HPG axis, a conclusion that is in accordance with our earlier analysis of AR113Q mice (45). Alternatively, the HPG axis may be particularly sensitive to altered regulation of AR target genes by the SUMOylation-deficient AR, perhaps due to tissue-specific differences in AR SUMOylation. This latter view is consistent with the androgen-insensitivity phenotype of patients with perturbed AR SUMOylation due to germline mutations in the first SUMOylation motif of AR (46).
Prior studies have suggested a modulatory role for SUMO in models of SBMA. Disrupting overall SUMOylation worsens the SBMA phenotype in a Drosophila model (47), and enhancing SUMOylation reduces polyQ AR aggregates, which are thought to reflect protein unfolding and the process that underlies proteotoxicity (31). We suspect that our results are not inconsistent with these findings. The Drosophila study used an amino-terminal fragment of the AR that lacks the SUMOylation sites and ligand-binding domain to define effects from global impairment of the SUMO pathway. This contrasts with the specific modulation of AR SUMOylation reported here. Similarly, the inhibitory effects of AR SUMOylation on aggregation require a high stoichiometry of modification. Thus, in cell culture, introducing the KRKR mutations into the AR without inducing high levels of SUMOylation does not modulate polyQ AR aggregation (31), a finding similar to our observations in vivo (Figure 7C).
Select point mutations in polyQ proteins exert varying influence on pathogenesis by modifying protein localization, function, or stability. In spinocerebellar ataxia type 1 (SCA1), disease is ameliorated by diminishing association of polyQ ataxin-1 with the transrepressor Capicua through the S776A mutation or by preventing nuclear transport through nuclear localization sequence disruption (48, 49). Consistent with these studies, phosphomimetic S776D ataxin-1 recapitulates toxicity, and pathogenic S776 phosphorylation and subsequent nuclear transport are prevented by association with 14-3-3 (50, 51). In SCA7, point mutations in the NES signal of polyQ ataxin-7 reduce nuclear export and exacerbate toxicity (52). Phosphomimetic mutations at Ser13 and Ser16 in polyQ huntingtin rescue disease and aggregation, while phospho-null mutations iterate pathogenesis (53). In this regard, we demonstrate ameliorative effects of KRKR point mutations in the polyQ AR that modulate AR function without modifying intracellular localization or aggregation. This new disease model advances our understanding of SBMA pathogenesis by demonstrating beneficial effects of enhancing function of the polyQ AR and revealing key features of AR SUMO modification that may be exploited for therapeutic targeting.
-
Methods
Antibodies. Primary antibodies used for Western blot analysis were against AR (N-20, Santa Cruz Biotechnology Inc.), Hsp90 (H-114, Santa Cruz Biotechnology Inc.), and HA (HA-11, Covance). Primary antibodies for immunofluorescence were against AR (N-20, Santa Cruz Biotechnology Inc.), type I/slow-twitch heavy-chain myosin (A4.840, DSHB, University of Iowa, Iowa City, Iowa, USA), and type II/fast-twitch heavy-chain myosin (Ab-2, Thermo Scientific). Horseradish peroxidase–conjugated secondary antibodies used for Western blot analysis were from Bio-Rad. Alexa Fluor 555, Alexa Fluor 488, and wheat germ agglutinin Alexa Fluor 488 conjugate antibodies used for immunofluorescence were from Invitrogen. FITC-conjugated donkey anti-mouse IgM was from Jackson ImmunoResearch. For IP experiments, the anti-AR antibody PG-21 (Millipore) was used, and pulldown was performed with protein A–agarose beads (Santa Cruz Biotechnology Inc.).
Cell culture, Western blot, and IP. PC12 cells expressing tetracycline-regulated AR10Q and AR112Q have been described previously (54). Tet-ON AR111Q-KRKR cells were generated by stable transfection of pTRE-AR(111-CAG)-KRKR, as described earlier (55). The construct was generated by excising a portion of p5HB-hAR24Q-KRKR containing the K385R and K518R mutations with NruI/Tth111I and cloning the fragment into pTRE-AR(111-CAG). The codons for K385 and K518 were AAG and AAA, respectively, and were mutated to codons for arginine AGA and CGA, respectively. Both K-to-R mutations and the CAG repeat tract-length were verified by sequencing. Induction of AR expression in AR10Q, AR112Q, and AR111Q-KRKR cells to equivalent protein levels was performed with 50 ng/ml, 3 ng/ml, and 500 ng/ml doxycycline (Clontech), respectively, in the presence of 10 nM R1881 or ethanol vehicle for 24 hours unless otherwise indicated. Cells were grown and passaged on poly-d-lysine–coated flasks in high-glucose phenol-red–free DMEM (Invitrogen) containing 10% charcoal-stripped horse serum (Invitrogen), 5% charcoal-stripped FBS (Atlantic Biologicals), 100 units/ml penicillin/streptomycin (Invitrogen), 200 μg/ml hygromycin B (Invitrogen), and 100 μg/ml G418 (Invitrogen) at 37°C in 10% CO2. For Western blot analysis, cells were washed twice and collected by cell scraper in PBS, then lysed in RIPA buffer supplemented with Halt phosphatase inhibitors (Thermo Scientific), complete protease inhibitors (Roche), and 20 mM N-ethylmaleimide (Sigma-Aldrich) to irreversibly inhibit SUMO proteases. Whole-cell lysates were preclarified by centrifugation at 15,000 g for 15 minutes at 4°C. Protein concentrations were determined by modified protein assay (Bio-Rad). After boiling at 100°C for 5 minutes in loading buffer, all protein samples were resolved on 7% or 10% SDS-PAGE gels, transferred to nitrocellulose membranes on a semi-dry transfer apparatus (Bio-Rad), and probed by the indicated antibodies. Detection was performed by chemiluminescence. For IP, 1500 μg of protein lysate from mouse tissues in 500 μl of RIPA buffer was incubated with 10 μg of PG-21 antibody overnight at 4°C. Protein A–agarose beads were prewashed 3 times in RIPA, then incubated in IP samples for 1 hour at 4°C. Bead-antibody complexes were then centrifuged in filtered spin columns (Thermo Scientific), washed 4 times in lysis buffer, and eluted by boiling in loading buffer for 5 minutes at 100°C. Testis lysates were obtained by homogenization and centrifugation at 13,000 g for 10 minutes at 4°C. Protein concentrations were determined as described above, and lysates were resolved on a 7.5% SDS-PAGE gel.
AR ubiquitination and SUMOylation studies. For analysis of ubiquitination, PC12 cells expressing AR112Q or AR111Q-KRKR were plated at 4 × 106 cells in a poly-d-lysine–coated 60-mm dish (BD Falcon) and transiently transfected with plasmids encoding hCHIP (1.5 μg) and HA-His-ubiquitin (4.5 μg) using Lipofectamine 2000 (Life Technologies). Cells were treated with 1 μg/ml doxycycline and 10 nM R1881 and then with 1 μM YM-1 (last 24 hours) and 10 μM MG132 (last 16 hours). 48 hours after transfection, cell pellets were resuspended in IP lysis/wash buffer (0.025 M Tris, 0.15 M NaCl, 0.001 M EDTA, 1% NP-40, 5% glycerol; pH 7.4) containing 1 tablet/10 ml of complete Mini-Protease Inhibitor Mix (Roche) and 5 mM N-ethylmaleimide. Samples were sonicated and centrifuged at 13,000 g for 10 minutes at 4°C. 400 μg of protein lysate was incubated on a rotator overnight at 4°C with 2 μg of anti-AR antibody (Santa Cruz Biotechnology Inc.) or nonimmune rabbit IgG. 20 μl of prewashed protein A–agarose beads (Santa Cruz Biotechnology Inc.) was added, and samples were incubated for 1 additional hour at 4°C. Protein-antibody-bead complexes were washed 6 times in IP lysis/wash buffer in filtered spin columns (Thermo Scientific), and proteins were eluted by boiling in SDS-loading buffer for 5 minutes at 100°C. A similar protocol was followed for analysis on SUMOylation, except that cells were transfected with plasmid encoding HA-SUMO3. Lysates were collected 24 hours after the addition of 1 μg/ml doxycycline and 10 nM R1881 in IP lysis/wash buffer containing 20 mM N-ethylmaleimide.
Generation of knockin mice. Knockin mice with AR allele containing 113 CAG repeats were derived using gene targeting as described previously (56). To generate AR113Q-KRKR mice, this strategy was modified by using the previously described AR113Q-targeting vector as a template. Briefly, a portion of p5HB-hAR24Q-KRKR containing the K385R and K518R mutations was excised as an NruI/XhoI fragment and cloned into pLP1-AR113Q. The codons for K385 and K518 were AAG and AAA, respectively, and were mutated to codons for arginine AGA and CGA, respectively. Both K-to-R mutations and the CAG repeat tract-length was verified by sequencing. The modified targeting construct containing amino acids 31–484 of human AR exon 1 and K385,518R mutations replaced mouse AR exon 1 by homologous recombination in JM8.F6 ES cells (57). Correct targeting of the AR113Q-KRKR allele was verified through Southern blot analysis using 5′ and 3′ probes falling outside of the targeted construct (Figure 2A). Three clones of correctly targeted ES cells were selected by neomycin resistance, karyotyped to verify euploidy, transiently transfected with Cre recombinase to excise the floxed neomycin resistance cassette, and injected into C57BL/6J blastocysts obtained from mating C57BL/6-BrdCr-Hsd-Tyrc females with C57BL/6-BrdCrHsd-Tyrc males. These blastocysts were then implanted in pseudopregnant C57BL/6J females. All ES cell procedures were performed by the University of Michigan Transgenic Animal Model Core.
Mouse breeding and care. ES cell implantations yielded 8 chimeric males in total, which were then mated with albino C57BL/6J females (Jackson Laboratory) to sire AR113Q-KRKR F1 female mice heterozygous for the targeted allele. Germline transmission of the targeted AR allele was indicated by black coat color and confirmed by PCR genotyping. These females were bred to C57BL/6J WT males to generate AR113Q-KRKR males for use in this study and AR113Q-KRKR heterozygous females to maintain the line. Mice were group housed in a specific pathogen–free (SPF) facility and provided with chow and water ad libitum. Genotypes were verified by PCR of DNA harvested from tail biopsies obtained shortly after weaning, using the forward primer 5′-CCAGAATCTGTTCCAGAGCGTG-3′ and the reverse primers 5′-TGTTCCCCTGGACTCAGATG-3′ and 5′-GCACTCCAGGGCCGACTGCG-3′ in a 2:2:1 ratio, respectively.
Mouse phenotyping and tissue collection. Every 2 weeks beginning at 8 weeks of age, male mice were weighed and forelimb strength was measured by grip strength meter (Columbus Instruments). Following acclimation to the instrument, mice were allowed to grip the triangular pull bar with forelimbs only and were then pulled backward horizontally; the average peak tension value from 5 consecutive grip trials was reported as the grip strength for each mouse. Disease onset was defined as the age at which grip strength was 10% less than the mean of age-matched controls. Skeletal muscles (quadriceps, gastrocnemius, soleus, TA, LABC), spinal cord, testis, and seminal vesicles were collected from adult males 13 to 15 weeks old and weighed or flash frozen in liquid nitrogen for biochemical analysis or mounted on OCT blocks for cryosectioning. Epididymes were also collected at this time, minced in 4 ml of PBS, incubated for 10 minutes at room temperature with gentle agitation, centrifuged for 1 minute at 500 g, and applied to a hemacytometer to determine sperm counts. For tissue analyses, quadriceps muscle and lumbar spinal cord were minced, lysed by agitation at 4°C for 1 hour, and homogenized in RIPA buffer containing phosphatase and protease inhibitors. Tissue lysates were then clarified at 13,000 g for 10 minutes at 4°C. Protein concentrations were determined by modified protein assay (Bio-Rad).
Measurement of serum hormone levels. Serum was collected from mice at 13 to 15 weeks by cardiac puncture, and testosterone and LH levels were determined by radioimmunoassay (RIA) and sandwich assay, respectively, by the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core Facility (Charlottesville, Virginia, USA). The sensitivities of the testosterone RIA and LH assay were 10 ng/dl and 0.07 ng/dl, respectively.
Exercise studies. To analyze exercise capacity, a protocol for treadmill running was used as previously described (58). First, mice underwent 2 days of training on an Exer3/6 treadmill (Columbus Instruments), with day 1 consisting of 5 minutes of running at 8 m/min and day 2 consisting of 5 minutes of running at 8 m/min followed by 5 minutes at 10 m/min. On day 3, mice ran using a graded protocol consisting of 10 m/min for 40 minutes, then increasing speed by increments of 1 m/min every 10 minutes for a total of 30 minutes, and followed by increasing speed by increments of 1 m/min every 5 minutes until mice were exhausted. Exhaustion was defined as mice making no attempt to exercise for 5 seconds. Assessment of endurance in castrated AR113Q-KRKR males was performed after orchiectomy at 6 weeks of age as previously described (36). Briefly, following anesthetization with isoflurane, the pelvic area was shaved and opened bilaterally with 1-cm incisions to reveal testes for removal, then closed with suture material.
Immunofluorescence analysis. PC12 cells were washed in PBS and fixed in 4% paraformaldehyde for 30 minutes at room temperature. For quadriceps and soleus muscles, cryosections were prepared using a Cryocut 1800 cryostat (Leica) at 10 μm. Staining was performed using the indicated antibodies, mounted using Vectashield medium (Vector Laboratories), and sealed with nail polish. We scored AR translocation from cytoplasm to nucleus as described previously (59, 60), using a value of 4 for nuclear fluorescence much greater than cytoplasmic fluorescence, 3 for nuclear fluorescence greater than cytoplasmic fluorescence, 2 for nuclear fluorescence equal to cytoplasmic fluorescence, 1 for nuclear fluorescence less than cytoplasmic fluorescence, and 0 for nuclear fluorescence much less than cytoplasmic fluorescence. The reported scores represent data from 50 cells per time point. To distinguish between type I and type II fibers, FITC-conjugated donkey anti-mouse IgM was used against A4.840 and Alexa Fluor 555 anti-mouse IgG was used against Ab-2. Fluorescence images were acquired using a Zeiss Axio Imager microscope. To calculate percentage of AR aggregation, AR-positive nuclear puncta were manually counted and divided by the total number of nuclei as quantified by ImageJ (NIH). To calculate fiber type size, the cross-sectional area of each fiber within a ×40 field was quantified in triplicate using ImageJ, and fiber type was discriminated by antibody stain.
Orchiectomy. Adult male mice underwent orchiectomy following induction of anesthesia by 5% isoflurane and maintenance by 1.5% to 2% isoflurane. The abdomen was cleaned and shaved, and a 5-mm incision was made at the level of the hind legs. The vas deferens and spermatic cord were visualized, separated, and ligated with nonabsorbable silk sutures. The testes were removed, and the incision was closed with absorbable surgical suture. Carpofen was given daily at 5 μg/g body weight by s.c. injection for 3 days after surgery. Three weeks after surgery, mice were treated with 200 μg testosterone propionate (Sigma-Aldrich) or sesame oil by i.p. injection. Tissue was harvested 16 hours after injection and stored in liquid nitrogen.
Gene-expression analysis. Total RNA was extracted from PC12 cells and from testes, prostate, quadriceps, and soleus muscles from mice with Trizol (Sigma-Aldrich). The RNA samples were then reverse transcribed with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s recommendations. qPCR was carried out using FastStart TaqMan Probe Master Mix (Roche) on software supplied with a 7500 Real-Time PCR SDS System (Applied Biosystems) and using gene-specific primers with FAM-labeled probes from Applied Biosystems (Mouse: Ar, Mm00442688_m1; Chrna1, Mm00431627_m1; Errfi1, Mm00505292_m1; Fkbp5, Mm00487401_m1; Hsd3b1, Mm00476184_g1; Igf1r, Mm00802831_m1; Irs2, Mm03038438_m1; Myod1, Mm00499518_m1; Myog, Mm00446194_m1; Nkx3-1, Mm00440479_m1; Pbsn, Mm00444381_m1; Spdef, Mm00600221_m1; Steap4, Mm00475405_m1; Wipi1, Mm00461219_m1; Zbtb10, Mm01281740_m1. Rat: Baiap2, Rn00589411_m1; Chst1, Rn01484520_m1; Ppard, Rn00565707_m1). Relative expression levels were calculated by normalizing to the expression of 18S rRNA. For GeneChip microarray analysis, cDNA samples from PC12 cells were applied to an Affymetrix Mouse Gene ST 2.1 Plate by the University of Michigan MicroArray Core. For RNA-Seq, cDNA was prepared into sequencer-ready fragment libraries using the Illumina TruSeq mRNA-Seq Sample Prep Kit according to the manufacturer’s recommended protocols and sequenced on the Illumina HiSeq 2000 using the manufacturer’s recommended protocols for paired-end, 100 nt sequencing. Samples were multiplexed into pools of 6 samples each, and each pool was sequenced on 1 lane of the sequencer. All original microarray and RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus (microarray data, GEO GSE50383; RNA-Seq data, GSE60691).
Statistics. To determine statistical significance, data sets were analyzed using unpaired 2-tailed Student’s t test, ANOVA with Newman-Keuls multiple comparison test, or Mantel-Cox log-rank test in Prism 6 (GraphPad). Differences between means were defined as significant at P < 0.05.
For GeneChip analysis, probe data were inspected for quality control by signal density distribution and normalized unscaled SEM estimates. Relative expression values were calculated using a robust multiarray average (RMA), with oligo and limma packages of bioconductor implemented in the R statistical environment; P values were adjusted for multiple comparisons using FDR as described previously (61–64). Expression values were also inspected for quality control by principal components analysis. Genes were considered significantly responsive to androgen if they demonstrated a 1.5-fold or greater activation or repression in the R1881-treated samples compared with controls and an adjusted P value of 0.05 or less. Genes selected in this manner and relevant information (probe set ID, statistics, expression values, gene name, symbol, and links to online databases) were output into Excel for ranking by fold change values and determining the numbers used for Venn diagrams.
For RNA-Seq, the software package Tuxedo Suite was used for alignment, differential expression analysis, and postanalysis diagnostics (65–67). The reference transcriptome (UCSC mm10) (http://genome.ucsc.edu/) was used for alignment. Expression quantitation and differential expression analysis were performed using CuffDiff (Cufflinks version 2.1.1). Genes and transcripts were identified as differentially expressed based on 3 criteria: test status = OK, FDR <0.05, and fold change ≥ 1.5. Analysis of differentially expressed genes to identify significantly enriched functional categories was performed using DAVID. The heat map was plotted in R using the gplots and RColorBrewer packages.
Study approval. The University of Michigan Committee on Use and Care of Animals approved all procedures involving mice in compliance with the Guide for the Care and Use of Laboratory Animals (8th ed. National Academies Press. Revised 2011).
-
Acknowledgments
We thank Susan Brooks for expertise and technical advice in fiber typing skeletal muscle, Steve Whitesall for training and assistance in performing treadmill exercise studies, Craig Johnson for microarray analysis, and Amanda Wong for experimental assistance. We thank Thomas Saunders and colleagues for assistance with the generation of gene-targeted mice through the University of Michigan Transgenic Animal Model Core and Robert Lyons and the University of Michigan DNA Sequencing Core for assistance with RNA-Seq. This work was supported by grants from the NIH (F31 NS076189 to J.P. Chua; R01 NS055746 and R03 NS084006 to A.P. Lieberman; R01 NCI144032 to D.M. Robins; R01 NS032214 to D.E. Merry).
Address correspondence to: Andrew Lieberman, Department of Pathology, University of Michigan Medical School, 3510 MSRB1, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0605, USA. Phone: 734.647.4624; E-mail: liebermn@umich.edu.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2015;125(2):831–845. doi:10.1172/JCI73214.
Heather L. Montie’s present address is: Department of Bio-Medical Sciences, Philadelphia College of Osteopathic Medicine, Philadelphia, Pennsylvania, USA.
See the related article at Fighting polyglutamine disease by wrestling with SUMO.
-
References
- Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247.
- Kennedy WR, Alter M, Sung JH. Progressive proximal spinal and bulbar muscular atrophy of late onset. A sex-linked recessive trait. Neurology. 1968;18(7):671–680.
- Katsuno M, et al. Reversible disruption of dynactin 1-mediated retrograde axonal transport in polyglutamine-induced motor neuron degeneration. J Neurosci. 2006;26(47):12106–12117.
- Sperfeld AD, et al. X-linked bulbospinal neuronopathy: Kennedy disease. Arch Neurol. 2002;59(12):1921–1926.View this article via: PubMed Google Scholar
- La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352(6330):77–79.
- Chamberlain NL, Driver ED, Miesfeld RL. The length and location of CAG trinucleotide repeats in the androgen receptor N-terminal domain affect transactivation function. Nucleic Acids Res. 1994;22(15):3181–3186.
- Irvine RA, Ma H, Yu MC, Ross RK, Stallcup MR, Coetzee GA. Inhibition of p160-mediated coactivation with increasing androgen receptor polyglutamine length. Hum Mol Genet. 2000;9(2):267–274.
- Kazemi-Esfarjani P, Trifiro MA, Pinsky L. Evidence for a repressive function of the long polyglutamine tract in the human androgen receptor: possible pathogenetic relevance for the (CAG)n-expanded neuronopathies. Hum Mol Genet. 1995;4(4):523–527.
- Mhatre AN, et al. Reduced transcriptional regulatory competence of the androgen receptor in X-linked spinal and bulbar muscular atrophy. Nat Genet. 1993;5(2):184–188.
- Lieberman AP, Harmison G, Strand AD, Olson JM, Fischbeck KH. Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum Mol Genet. 2002;11(17):1967–1976.
- Morfini G, Pigino G, Szebenyi G, You Y, Pollema S, Brady ST. JNK mediates pathogenic effects of polyglutamine-expanded androgen receptor on fast axonal transport. Nat Neurosci. 2006;9(7):907–916.
- Szebenyi G, et al. Neuropathogenic forms of huntingtin and androgen receptor inhibit fast axonal transport. Neuron. 2003;40(1):41–52.
- Ranganathan S, Harmison GG, Meyertholen K, Pennuto M, Burnett BG, Fischbeck KH. Mitochondrial abnormalities in spinal and bulbar muscular atrophy. Hum Mol Genet. 2009;18(1):27–42.View this article via: PubMed Google Scholar
- McCampbell A, et al. CREB-binding protein sequestration by expanded polyglutamine. Hum Mol Genet. 2000;9(14):2197–2202.
- Kemp MQ, et al. Impaired motoneuronal retrograde transport in two models of SBMA implicates two sites of androgen action. Hum Mol Genet. 2011;20(22):4475–4490.
- Lieberman AP, et al. Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy. Cell Rep. 2014;7(3):774–784.
- Cortes CJ, et al. Muscle expression of mutant androgen receptor accounts for systemic and motor neuron disease phenotypes in spinal and bulbar muscular atrophy. Neuron. 2014;82(2):295–307.
- Iniguez-Lluhi JA, Pearce D. A common motif within the negative regulatory regions of multiple factors inhibits their transcriptional synergy. Mol Cell Biol. 2000;20(16):6040–6050.
- Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc Natl Acad Sci U S A. 2000;97(26):14145–14150.
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11(12):861–871.
- Kaikkonen S, et al. SUMO-specific protein 1 (SENP1) reverses the hormone-augmented SUMOylation of androgen receptor and modulates gene responses in prostate cancer cells. Mol Endocrinol. 2009;23(3):292–307.
- Chupreta S, Holmstrom S, Iniguez-Lluhi JA. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol Cell Biol. 2005;25(10):4272–4282.
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101(40):14373–14378.
- Holmstrom SR, Chupreta S, Yick-Lun So A, Iniguez-Lluhi JA. SUMO-mediated of glucocorticoid receptor synergistic activity depends on stable assembly at the promoter but not on DAXX. Mol Endocrinol. 2008;22(9):2061–2075.
- Rytinki M, Kaikkonen S, Sutinen P, Paakinaho V, Rahkama V, Palvimo JJ. Dynamic SUMOylation is linked to the activity cycles of androgen receptor in the cell nucleus. Mol Cell Biol. 2012;32(20):4195–4205.
- Steffan JS, et al. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304(5667):100–104.
- O’Rourke JG, et al. SUMO-2 and PIAS1 modulate insoluble mutant huntingtin protein accumulation. Cell Rep. 2013;4(2):362–375.
- Dorval V, Fraser PE. SUMO on the road to neurodegeneration. Biochim Biophys Acta. 2007;1773(6):694–706.
- Mo K, et al. Microarray analysis of gene expression by skeletal muscle of three mouse models of Kennedy disease/spinal bulbar muscular atrophy. PLoS One. 2010;5(9):e12922.
- Nedelsky NB, et al. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 2010;67(6):936–952.
- Mukherjee S, Thomas M, Dadgar N, Lieberman AP, Iniguez-Lluhi JA. Small ubiquitin-like modifier (SUMO) modification of the androgen receptor attenuates polyglutamine-mediated aggregation. J Biol Chem. 2009;284(32):21296–21306.
- Wang AM, et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat Chem Biol. 2013;9(2):112–118.View this article via: PubMed Google Scholar
- Sutinen P, Malinen M, Heikkinen S, Palvimo JJ. SUMOylation modulates the transcriptional activity of androgen receptor in a target gene and pathway selective manner. Nucleic Acids Res. 2014;42(13):8310–8319.
- O’Shaughnessy PJ, Johnston H, Willerton L, Baker PJ. Failure of normal adult Leydig cell development in androgen-receptor-deficient mice. J Cell Sci. 2002;115(pt 17):3491–3496.View this article via: PubMed Google Scholar
- Kim JY, Park KD, Kim SM, Sunwoo IN. Decremental responses to repetitive nerve stimulation in x-linked bulbospinal muscular atrophy. J Clin Neurol. 2013;9(1):32–35.
- Yu Z, et al. Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. J Clin Invest. 2006;116(10):2663–2672.
- Costill DL, Daniels J, Evans W, Fink W, Krahenbuhl G, Saltin B. Skeletal muscle enzymes and fiber composition in male and female track athletes. J Appl Physiol. 1976;40(2):149–154.View this article via: PubMed Google Scholar
- Fink WJ, Costill DL, Pollock ML. Submaximal and maximal working capacity of elite distance runners. Part II. Muscle fiber composition and enzyme activities. Ann N Y Acad Sci. 1977;301:323–327.
- Klover P, Chen W, Zhu BM, Hennighausen L. Skeletal muscle growth and fiber composition in mice are regulated through the transcription factors STAT5a/b: linking growth hormone to the androgen receptor. FASEB J. 2009;23(9):3140–3148.
- Sher J, Cardasis C. Skeletal muscle fiber types in the adult mouse. Acta Neurol Scand. 1976;54(1):45–56.
- Jiang M, et al. Androgen-responsive gene database: integrated knowledge on androgen-responsive genes. Mol Endocrinol. 2009;23(11):1927–1933.
- Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91(4):1447–1531.
- Nishida T, Yasuda H. PIAS1 and PIASxalpha function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J Biol Chem. 2002;277(44):41311–41317.
- Kaikkonin S, et al. SUMO-Specific Protease 1 (SENP1) reverses the hormone-augmented SUMOylation of androgen receptor and modulates gene responses in prostate cancer cells. Mol Endocriol. 2009;23(3):292–307.
- Yu Z, et al. Abnormalities of germ cell maturation and sertoli cell cytoskeleton in androgen receptor 113 CAG knock-in mice reveal toxic effects of the mutant protein. Am J Pathol. 2006;168(1):195–204.
- Mukherjee S, Cruz-Rodriguez O, Bolton E, Iniguez-Lluhi JA. The in vivo role of androgen receptor SUMOylation as revealed by androgen insensitivity syndrome and prostate cancer mutations targeting the proline/glycine residues of synery control motifs. J Biol Chem. 2012;287(37):31195–31206.
- Chan HY, Warrick JM, Andriola I, Merry D, Bonini NM. Genetic modulation of polyglutamine toxicity by protein conjugation pathways in Drosophila. Hum Mol Genet. 2002;11(23):2895–2904.
- Lam YC, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127(7):1335–1347.
- Klement IA, et al. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95(1):41–53.
- Duvick L, et al. SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010;67(6):929–935.
- Lai S, O’Callaghan B, Zoghbi HY, Orr HT. 14-3-3 Binding to ataxin-1(ATXN1) regulates its dephosphorylation at Ser-776 and transport to the nucleus. J Biol Chem. 2011;286(40):34606–34616.
- Taylor J, et al. Ataxin-7 can export from the nucleus via a conserved exportin-dependent signal. J Biol Chem. 2006;281(5):2730–2739.View this article via: PubMed Google Scholar
- Gu X, et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64(6):828–840.
- Walcott JL, Merry DE. Ligand promotes intranuclear inclusions in a novel cell model of spinal and bulbar muscular atrophy. J Biol Chem. 2002;277(52):50855–50859.
- Montie HL, Pestell RG, Merry DE. SIRT1 modulates aggregation and toxicity through deacetylation of the androgen receptor in cell models of SBMA. J Neurosci. 2011;31(48):17425–17436.
- Albertelli MA, Scheller A, Brogley M, Robins DM. Replacing the mouse androgen receptor with human alleles demonstrates glutamine tract length-dependent effects on physiology and tumorigenesis in mice. Mol Endocrinol. 2006;20(6):1248–1260.
- Pettitt SJ, et al. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods. 2009;6(7):493–495.
- He C, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481(7382):511–515.
- Thomas M, et al. The unfolded protein response modulates toxicity of the expanded glutamine androgen receptor. J Biol Chem. 2005;280(22):21264–21271.
- Thomas M, Harrell JM, Morishima Y, Peng HM, Pratt WB, Lieberman AP. Pharmacologic and genetic inhibition of hsp90-dependent trafficking reduces aggregation and promotes degradation of the expanded glutamine androgen receptor without stress protein induction. Hum Mol Genet. 2006;15(11):1876–1883.
- Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264.
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300.
- Ritchie ME, et al. Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics. 2006;7:261.
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3.View this article via: PubMed Google Scholar
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25.
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53.View this article via: PubMed Google Scholar
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111.
-
Version history
- Version 1 (January 20, 2015): No description
- Version 2 (February 2, 2015): No description



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)










