Advertisement
Review Series Free access | 10.1172/JCI67229
Detection of oncogenic IDH1 mutations using magnetic resonance spectroscopy of 2-hydroxyglutarate
Ovidiu C. Andronesi,1 Otto Rapalino,2 Elizabeth Gerstner,3,4 Andrew Chi,3,4 Tracy T. Batchelor,3,4,5 Dan P. Cahill,6 A. Gregory Sorensen,7 and Bruce R. Rosen1
1Athinoula A. Martinos Center for Biomedical Imaging, 2Department of Radiology, 3Department of Neurology, 4Division of Neuro-Oncology, 5Department of Radiation Oncology, and 6Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA. 7Siemens Healthcare USA, Malvern, Pennsylvania, USA.
Address correspondence to: Ovidiu C. Andronesi, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Thirteenth Street, Building 149, Suite 2301, Boston, Massachusetts, USA. Phone: 617.643.6864; Fax: 617.726.7422; E-mail: ovidiu@nmr.mgh.harvard.edu.
Find articles by Andronesi, O. in: PubMed | Google Scholar
1Athinoula A. Martinos Center for Biomedical Imaging, 2Department of Radiology, 3Department of Neurology, 4Division of Neuro-Oncology, 5Department of Radiation Oncology, and 6Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA. 7Siemens Healthcare USA, Malvern, Pennsylvania, USA.
Address correspondence to: Ovidiu C. Andronesi, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Thirteenth Street, Building 149, Suite 2301, Boston, Massachusetts, USA. Phone: 617.643.6864; Fax: 617.726.7422; E-mail: ovidiu@nmr.mgh.harvard.edu.
Find articles by Rapalino, O. in: PubMed | Google Scholar
1Athinoula A. Martinos Center for Biomedical Imaging, 2Department of Radiology, 3Department of Neurology, 4Division of Neuro-Oncology, 5Department of Radiation Oncology, and 6Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA. 7Siemens Healthcare USA, Malvern, Pennsylvania, USA.
Address correspondence to: Ovidiu C. Andronesi, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Thirteenth Street, Building 149, Suite 2301, Boston, Massachusetts, USA. Phone: 617.643.6864; Fax: 617.726.7422; E-mail: ovidiu@nmr.mgh.harvard.edu.
Find articles by Gerstner, E. in: PubMed | Google Scholar
1Athinoula A. Martinos Center for Biomedical Imaging, 2Department of Radiology, 3Department of Neurology, 4Division of Neuro-Oncology, 5Department of Radiation Oncology, and 6Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA. 7Siemens Healthcare USA, Malvern, Pennsylvania, USA.
Address correspondence to: Ovidiu C. Andronesi, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Thirteenth Street, Building 149, Suite 2301, Boston, Massachusetts, USA. Phone: 617.643.6864; Fax: 617.726.7422; E-mail: ovidiu@nmr.mgh.harvard.edu.
Find articles by Chi, A. in: PubMed | Google Scholar
1Athinoula A. Martinos Center for Biomedical Imaging, 2Department of Radiology, 3Department of Neurology, 4Division of Neuro-Oncology, 5Department of Radiation Oncology, and 6Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA. 7Siemens Healthcare USA, Malvern, Pennsylvania, USA.
Address correspondence to: Ovidiu C. Andronesi, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Thirteenth Street, Building 149, Suite 2301, Boston, Massachusetts, USA. Phone: 617.643.6864; Fax: 617.726.7422; E-mail: ovidiu@nmr.mgh.harvard.edu.
Find articles by Batchelor, T. in: PubMed | Google Scholar
1Athinoula A. Martinos Center for Biomedical Imaging, 2Department of Radiology, 3Department of Neurology, 4Division of Neuro-Oncology, 5Department of Radiation Oncology, and 6Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA. 7Siemens Healthcare USA, Malvern, Pennsylvania, USA.
Address correspondence to: Ovidiu C. Andronesi, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Thirteenth Street, Building 149, Suite 2301, Boston, Massachusetts, USA. Phone: 617.643.6864; Fax: 617.726.7422; E-mail: ovidiu@nmr.mgh.harvard.edu.
Find articles by Cahill, D. in: PubMed | Google Scholar
1Athinoula A. Martinos Center for Biomedical Imaging, 2Department of Radiology, 3Department of Neurology, 4Division of Neuro-Oncology, 5Department of Radiation Oncology, and 6Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA. 7Siemens Healthcare USA, Malvern, Pennsylvania, USA.
Address correspondence to: Ovidiu C. Andronesi, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Thirteenth Street, Building 149, Suite 2301, Boston, Massachusetts, USA. Phone: 617.643.6864; Fax: 617.726.7422; E-mail: ovidiu@nmr.mgh.harvard.edu.
Find articles by Sorensen, A. in: PubMed | Google Scholar
1Athinoula A. Martinos Center for Biomedical Imaging, 2Department of Radiology, 3Department of Neurology, 4Division of Neuro-Oncology, 5Department of Radiation Oncology, and 6Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA. 7Siemens Healthcare USA, Malvern, Pennsylvania, USA.
Address correspondence to: Ovidiu C. Andronesi, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Thirteenth Street, Building 149, Suite 2301, Boston, Massachusetts, USA. Phone: 617.643.6864; Fax: 617.726.7422; E-mail: ovidiu@nmr.mgh.harvard.edu.
Find articles by Rosen, B. in: PubMed | Google Scholar
Published September 3, 2013 - More info
J Clin Invest. 2013;123(9):3659–3663. https://doi.org/10.1172/JCI67229.
© 2013 The American Society for Clinical Investigation
-
Abstract
The investigation of metabolic pathways disturbed in isocitrate dehydrogenase (IDH) mutant tumors revealed that the hallmark metabolic alteration is the production of D-2-hydroxyglutarate (D-2HG). The biological impact of D-2HG strongly suggests that high levels of this metabolite may play a central role in propagating downstream the effects of mutant IDH, leading to malignant transformation of cells. Hence, D-2HG may be an ideal biomarker for both diagnosing and monitoring treatment response targeting IDH mutations. Magnetic resonance spectroscopy (MRS) is well suited to the task of noninvasive D-2HG detection, and there has been much interest in developing such methods. Here, we review recent efforts to translate methodology using MRS to reliably measure in vivo D-2HG into clinical research.
-
Introduction
Recurrent heterozygous somatic mutations of the isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) genes were recently found by genome-wide sequencing to be highly frequent (50%–80%) in human grade II–IV gliomas (1, 2). IDH mutations are also often observed in several other cancers, including acute myeloid leukemia (3), central/periosteal chondrosarcoma and enchondroma (4), and intrahepatic cholangiocarcinoma (5). The identification of frequent IDH mutations in multiple cancers suggests that this pathway is involved in oncogenesis. Indeed, increasing evidence demonstrates that IDH mutations alter downstream epigenetic and genetic cellular signal transduction pathways in tumors (6, 7). In gliomas, IDH1 mutations appear to define a distinct clinical subset of tumors, as these patients have a 2- to 4-fold longer median survival compared with patients with wild-type IDH1 gliomas (8). IDH1 mutations are especially common in secondary glioblastoma (GBM) arising from lower-grade gliomas, arguing that these mutations are early driver events in this disease (9). Despite aggressive therapy with surgery, radiation, and cytotoxic chemotherapy, average survival of patients with GBM is less than 2 years, and less than 10% of patients survive 5 years or more (10).
The discovery of cancer-related IDH1 mutations has raised hopes that this pathway can be targeted for therapeutic benefit (11, 12). Methods that can rapidly and noninvasively identify patients for clinical trials and determine the pharmacodynamic effect of candidate agents in patients enrolled in trials are particularly important to guide and accelerate the translation of these treatments from bench to bedside. Magnetic resonance spectroscopy (MRS) can play an important role in clinical and translational research because IDH mutated tumor cells have such a distinct molecular phenotype (13, 14).
-
Biochemistry and metabolic alterations in IDH-mutated tumor cells
The family of IDH enzymes includes three isoforms: IDH1, which localizes in peroxisomes and cytoplasm, and IDH2 and IDH3, which localize in mitochondria as part of the tricarboxylic acid cycle (11). All three wild-type enzymes catalyze the oxidative decarboxylation of isocitrate to α-ketoglutarate (αKG), using the cofactor NADP+ (IDH1 and IDH2) or NAD+ (IDH3) as the electron acceptor. To date, only mutations of IDH1 and IDH2 have been identified in human cancers (11), and only one allele is mutated. In gliomas, about 90% of IDH mutations involve a substitution in IDH1 in which arginine 132 (R132) from the catalytic site is replaced by a histidine (IDH1 R132H), known as the canonical IDH1 mutation (8). A number of noncanonical mutations such as IDH1 R132C, IDH1 R132S, IDH1 R132L, and IDH1 R132G are less frequently present. Arginine R172 in IDH2 is the corresponding residue to R132 in IDH1, and the most common mutation is IDH2 R172K. In addition to IDH2 R172K, IDH2 R140Q has also been observed in acute myeloid leukemia. Although most IDH1 mutations occur at R132, a small number of mutations producing D-2-hydroxyglutarate (D-2HG) occur at R100, G97, and Y139 (15). However, only a single residue is mutated in either IDH1 or IDH2 in a given tumor.
IDH mutations result in a very high accumulation of the oncometabolite D-2HG in the range of 5- to 35-mM levels, which is 2–3 orders of magnitude higher than D-2HG levels in tumors with wild-type IDH or in healthy tissue (13). All IDH1 G97, R100, R132, and Y139 and IDH2 R140 and R172 mutations confer a neomorphic activity to the IDH1/2 enzymes, switching their activity toward the reduction of αKG to D-2HG, using NADPH as a cofactor (15). The gain of function conferred by these mutations is possible because in each tumor cell a copy of the wild-type allele exists to supply the αKG substrate and NADPH cofactor for the mutated allele.
A cause and effect relationship between IDH mutation and tumorigenesis is probable, and D-2HG appears to play a pivotal role as the relay agent. Evidence is mounting that high levels of D-2HG alter the biology of tumor cells toward malignancy by influencing the activity of enzymes critical for regulating the metabolic (14) and epigenetic state of cells (6, 7, 16–18). D-2HG may act as an oncometabolite via competitive inhibition of αKG-dependent dioxygenases (16). This includes inhibition of histone demethylases and 5-methlycytosine hydroxylases (e.g., TET2), leading to genome-wide alterations in histone and DNA hypermethylation as well as inhibition of hydroxylases, resulting in upregulation of HIF-1 (19). The effects of D-2HG have been shown to be reversible in leukemic transformation (18), which gives further evidence that treatments that lower D-2HG could be a valid therapeutic approach for IDH-mutant tumors. In addition to increased D-2HG, widespread metabolic disturbances of the cellular metabolome have been measured in cells with IDH mutations, including changes in amino acid concentration (increased levels of glycine, serine, threonine, among others, and decreased levels of aspartate and glutamate), N-acetylated amino acids (N-acetylaspartate, N-acetylserine, N-acetylthreonine), glutathione derivatives, choline metabolites, and TCA cycle intermediates (fumarate, malate) (14). These metabolic changes might be exploited for therapy. For example, IDH mutations cause a depletion of NADPH, which lowers the reductive capabilities of tumor cells (20) and perhaps makes them more susceptible to treatments that create free radicals (e.g., radiation) (21).
-
In vivo MRS of D-2HG in IDH mutant tumors
D-2HG may be an optimal biomarker for tumors with IDH mutations, as it ideally fulfills several important requirements: (a) there is virtually no normal D-2HG background — in cells without IDH mutations, D-2HG is produced as an error product of normal metabolism and is only present at trace levels; (b) 99% of tumors with IDH mutations have increased levels of D-2HG by several orders of magnitude; (c) the only other known cause of elevated 2HG is hydroxyglutaric aciduria (in this case, high L-2HG caused by a mutation in 2HG dehydrogenase), which is a rare inborn error of metabolism that presents with a different clinical phenotype and marked developmental anomalies in early childhood. Hence, tumors displaying increased levels of D-2HG are unlikely to represent false-positive cases for IDH mutations. Furthermore, this raises the possibility that D-2HG levels could also be used to quantify and predict the efficacy of drugs targeting mutant IDH1 for antitumor therapy (11, 15). In fact, it is hard to find a similar example of another tumor biomarker metabolite that is so well supported by the underlying biology.
The high levels of D-2HG observed in IDH1-mutant gliomas are amenable to detection by in vivo MRS. Given that the detection threshold of in vivo MRS is around 1 mM (1 μmol/g, wet tissue), D-2HG should be measurable only in situations in which it accumulates due to IDH1 mutations. Conversely, D-2HG is not expected to be detectable in tumors in which IDH1 is not mutated or in healthy tissues. In addition, ex vivo MRS measurements of intact biopsies (22) or extracts reach higher sensitivity 0.1–0.01 mM (0.1–0.01 μmol/g) and can be used as a cheaper and faster alternative to mass spectrometry.
However, unambiguous measurement of 2HG with proton MRS, especially in vivo, is challenging because its coupled five-spin system yields a complex spectral pattern (23) that overlaps with other metabolites that are normally present in the brain or may accumulate in cancer. The D-2HG signals from Hβ (1.91 ppm) and Hγ (2.24 ppm) protons are superimposed by glutamate, glutamine, and γ-aminobutyric acid (GABA), while Hα (4.02 ppm) signals are obscured by myoinositol, phosphocreatine, and lactate.
Spectral fitting of conventional 1H magnetic resonance spectra is likely to provide a high rate of false-positive results for D-2HG or may lead to under- or overestimation of D-2HG levels. Indeed, a false-positive rate of approximately 22% was observed by Pope et al. (24) when using conventional MRS and spectral fitting to detect D-2HG in glioma patients. This error rate is a result of the spectral decomposition of the fitting algorithm, which may not be unique, especially when dealing with the limited resolution of in vivo spectra acquired at low fields. This type of error is well documented for GABA, which poses similar problems (25). Indirect proof regarding the difficulty of D-2HG detection is provided by the fact that despite the high frequency of IDH1 mutations in glioma, high levels of D-2HG, and large number of MRS studies of brain tumors performed over the last 20 years (26–28), the existence of D-2HG was not reported in the cancer MRS literature until after it was discovered through genomic and ex vivo metabolomic analysis (1, 2, 13).
Recently, reliable detection of D-2HG using in vivo 1H MRS was demonstrated in glioma patients (29, 30). Andronesi et al. reported the unambiguous detection of D-2HG in mutant IDH1 glioma in vivo using 2D correlation spectroscopy (COSY) and J-difference spectroscopy (29). In 2D COSY the overlapping signals are resolved along a second orthogonal chemical shift dimension (31, 32), and in the case of D-2HG, the cross-peaks resulting from the scalar coupling of Hα-Hβ protons show up in a region that is free of the contribution of other metabolites in both healthy and wild-type tumors. While 2D COSY retains all the metabolites in the spectrum, J-difference spectroscopy (25, 33) takes the opposite approach instead by focusing on the metabolite of interest, such as D-2HG, and selectively applying a narrow-band radiofrequency pulse to selectively refocus the Hα-Hβ scalar coupling evolution, then removing the contribution of overlapping metabolites. In this case a 1D difference spectrum with the Hα signal of D-2HG is detected at 4.02 ppm. Both methods have strengths and weaknesses: 2D COSY has the highest resolving power to disentangle overlapping metabolites, but has less sensitivity and quantification is more complex; J-difference spectroscopy has increased sensitivity, and quantification is straightforward, but it is susceptible to subtraction errors.
In a study conducted on glioma patients, Choi et al. (30) detected and quantified D-2HG levels using spectral editing at long echo times (TEs) and J-difference spectroscopy. Spectral editing at long TEs takes advantage of the modulation of multiplets and line shapes due to scalar coupling evolution and achieves some degree of filtering based on this mechanism and difference in transverse relaxation. In the case of D-2HG, a TE of 97 ms maximizes the contribution of Hγ protons (2.24 ppm) against the background of glutamate and glutamine. This approach has the advantage of simplicity because it uses sequences existing on all MR scanners and relies on the fitting to separate D-2HG from glutamate, glutamine, and other metabolites at TE of 97 ms. Good spectral resolution, scanner stability, and absence of motion artifacts is required to reliably disentangle signals at 2.24 ppm. Detection based on spectral difference has been obtained based on the same principle outlined above. In their study, Choi et al were able to correctly identify all patients that have IDH mutations (15 of 30), without false-positive results. They also quantified levels of D-2HG based on the internal water reference signal, and in their patient population in vivo D-2HG was found to range between 2 and 9 mM.
In Table 1, a comparison is made among the published methods for D-2HG detection. Results selected from the literature are shown in Figure 1. Besides the approaches discussed thus far, other methods are available in the in vivo MRS armamentarium that could be perhaps explored for reliable detection of 2D-HG, such as multiple-quantum filtering sequences (34, 35) and a variety of 2D spectroscopic methods (36–39).
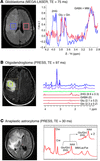 Figure 1
Figure 1In vivo D-2HG measurements: (A) J-difference spectroscopy with MEGA-LASER sequence in a patient with GBM with mutant IDH1. Adapted with permission from Science Translational Medicine (29). (B) Spectral editing with PRESS sequence of TE 97 ms (TE1: 32 ms, TE2: 65 ms) in a patient with mutant IDH1 oligodendroglioma. Adapted with permission from Nature Medicine (30). (C) Spectra acquired with PRESS sequence of TE 30 ms in a patient with mutant IDH1 anaplastic astrocytoma. Adapted with permission from Journal of Neuro-Oncology (24). Cho, choline; Cre, creatine; Gln, glutamine; Glu, glutamate; Lac, lactate; MM, macromolecules; NAA, N-acetyl-aspartate.
 Table 1
Table 1Summary of in vivo 1H MRS methods used in the literature for detection of D-2HG in patients with mutant IDH glioma
-
Caveats of in vivo MRS
A major determinant of in vivo MRS performance is signal localization. Most in vivo MRS sequences are based on point-resolved spectroscopy (PRESS) (40), employing radiofrequency pulses with limited bandwidth, which results in large chemical shift displacement error, and are affected by B1 field inhomogeneity, in particular for B0 fields above 1.5 T. This approach leads to signal loss via inaccurate localization and variability of flip angle, especially in J-difference spectra (41, 42). Improved localization and signal to noise may be obtained for multiplets of coupled spins using localized adiabatic spin echo refocusing (LASER) (43, 44), which minimizes chemical shift artifact and compensates for B1 inhomogeneity.
J-difference spectroscopy, multiple quantum filtering, and 2D methods require longer acquisition times, which makes them more susceptible to errors due to subject motion and hardware instabilities, causing drifts in the main magnetic field B0. In order to account for these factors, acquisition can be broken in several shorter time blocks with scanner adjustment in between and retrospectively discarding averages that were corrupted by motion (45). A more precise way for achieving this is to perform prospective real-time motion correction and dynamic shim (46, 47).
Absolute quantification of metabolites is important to assess in vivo enzymatic activity by MRS, but existing methods based on water reference have shortcomings when applied to tumors due to variations in water content (48). Relative quantification, typically employed in clinical exams, is also affected by confounding effects due to changes in the reference metabolite (49).
-
Ex vivo MRS of D-2HG in tumors with IDH mutations
The panoply of methods and ability of ex vivo MRS (50) to detect D-2HG in patient samples is far superior to in vivo MRS because the above list of limitations and artifacts is not of concern.
Metabolic profiling of intact tumor biopsies as small as 1 mg can be performed with high-resolution magic angle spinning (HRMAS) (51–53). HRMAS preserves the integrity of the samples that can be further analyzed with immunohistochemistry, genomics, or other metabolic profiling tools such as mass spectrometry. Detection of D-2HG in mutant IDH1 glioma was confirmed by ex vivo HRMAS experiments (29, 54, 55). In addition to D-2HG, ex vivo HRMAS studies can detect quantitative and qualitative changes for a large number of metabolites in IDH mutated tumors (54, 55).
However, as with any ex vivo assay, challenges involved with HRMAS include obtaining a representative tumor specimen, repeatability, and sample degradation. The main limitation of ex vivo measurements is, of course, the need of a biopsy, which in the case of brain tumors incurs risks and might not be repeatedly obtained with ease. In addition, the availability of high-resolution NMR spectrometers compared with MRI scanners is rare in most clinical sites, although tissue samples can be preserved frozen and shipped to a site with such equipment. An alternative to HRMAS of intact biopsies is the measurement of tumor extracts using perchloric acid. However, the extensive sample preparation required with this technique is prone to alter the metabolite content in tumor extracts.
-
Discussions and future implications
The example of IDH1 mutations is a perfect illustration of the rapid pace of progress brought to the medical sciences by the power and advances of modern technology: genome-wide sequencing, metabolomics, and imaging.
In vivo MRS has the unique ability to noninvasively probe IDH mutations by measuring the endogenously produced oncometabolite D-2HG. As an imaging-based technique, it has the benefit of posing minimal risk to the patients, can be performed repeatedly as many times as necessary, and can probe tumor heterogeneity without disturbing the internal milieu. To date, in vivo MRS is the only imaging method that is specific to IDH mutations — existing PET or SPECT radiotracers are not specific (56, 57), IDH-targeted agents for in vivo molecular imaging do not yet exist, and the prohibitive cost of radiotracers will likely limit their clinical development.
Caution must be exercised, however, when acquiring and analyzing in vivo MRS data. Reliable measurements of D-2HG require customized in vivo MRS sequences to avoid false-positive results. D-2HG has been measured by in vivo MRS in glioma patients, but the potential of in vivo MRS to probe IDH mutations remains to be demonstrated in other cancers. While in most tumors the levels of D-2HG seems to be high enough to grant detection by in vivo MRS, there may be a number of tumors in which levels could be around or below the 1-mM detection threshold, depending on the tumor stage or treatment manipulation. Such a gray zone needs to be further investigated to establish the limits or improve the method.
Other aspects are worth mentioning when comparing in vivo MRS with ex vivo assays. In vivo MRS may be faster and less expensive, as spectra can be read at the time of scan, spectra are available prior to biopsy, and no reagents are involved. In fact, clinicians are starting to include the MRS-derived information regarding the IDH status for neurosurgical planning (58). In particular, glioma IDH mutations are routinely diagnosed based on immunohistochemistry using antibodies targeted against IDH1 R132H (59), which miss around 15% of all IDH mutations. In vivo MRS has also been shown to detect those IDH mutations that are missed by immunohistochemistry (29, 30, 58). Moreover, gene sequencing, which is the ultimate diagnostic for IDH mutation, is not without problems. Sequencing may detect numerous SNPs, uncharacterized artifacts, and/or passenger alterations that have no effect on IDH enzyme activity (15). Also, sequencing is not available in most clinical institutions, and where available, results may be delayed by several weeks.
An alternative to D-2HG detection by in vivo MRS is D-2HG measurement in biofluids (plasma, urine, CSF) using gas/liquid-chromatography tandem mass spectrometry (GC-MS, LC-MS). This approach has been shown in acute myeloid leukemia patients (60), but to date it has failed in glioma patients (61).
Finally, as knowledge about cellular mechanisms continues to accumulate, the possibility of an antitumor treatment targeting IDH mutations may soon become reality (12). D-2HG may serve as a pharmacodynamic biomarker, providing information about whether drugs hit their target and allowing optimization of dosing. Hence, in vivo MRS may help optimize drug design and more quickly translate novel therapies to patients. This strategy is especially important in patients with mutant IDH glioma (which thus far represents the bulk of solid cancers with IDH mutations) and in situations in which nonimaging approaches are problematic (e.g., multiple biopsies in a longitudinal study are not feasible, tumors are heterogeneous, and detection of 2HG in biofluids has not been successful to date) (61).
By a fortunate combination of biological and technical factors that reinforce each other, in vivo MRS has a great potential to noninvasively genotype and monitor IDH mutations in cancer patients. Feasibility studies in glioma patients have shown that D-2HG measurements by in vivo MRS have a very high positive predictive value for IDH mutations. Further validation in other IDH cancers, investigation of reproducibility, repeatability, limits of detection, and improvements for longitudinal studies may be necessary, especially for clinical trials of new drugs targeting mutant IDH enzymes.
-
Acknowledgments
The MRS of IDH glioma patients was initiated by A.G. Sorensen while he was employed at Massachusetts General Hospital; A.G. Sorensen is currently employed by Siemens Healthcare USA. Discussions with Matthew G. vander Heiden were extremely valuable throughout the course of the study. O.C. Andronesi was supported by a KL2 Medical Research Investigator Training (MeRIT) award from Harvard Catalyst and by The Harvard Clinical and Translational Science Center (grant KL2 RR 025757). Additional funding was provided by the NIH for T.T. Batchelor (grants K24CA125440A and R01CA129371) and by shared instrumentation grants (S10RR013026, S10RR021110, and S10RR023401 from Martinos Center for Biomedical Imaging).
Address correspondence to: Ovidiu C. Andronesi, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Thirteenth Street, Building 149, Suite 2301, Boston, Massachusetts, USA. Phone: 617.643.6864; Fax: 617.726.7422; E-mail: ovidiu@nmr.mgh.harvard.edu.
-
Footnotes
Conflict of interest: Ovidiu C. Andronesi and A. Gregory Sorensen have applied for a patent for the 2D COSY-LASER method that was used for in vivo D-2HG detection (U.S. Patent Application Serial No. 13/237,799).
Reference information: J Clin Invest. 2013;123(9):3659–3663. doi:10.1172/JCI67229.
-
References
- Parsons DW, et al. An integrated genomic analysis of human glioblastoma Multiforme. Science. 2008;321(5897):1807–1812.
- Chin L, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068.
- Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066.
- Amary MF, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224(3):334–343.
- Borger DR, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17(1):72–79.
- Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478.
- Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483.
- Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773.
- Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–1153.
- http://www.cbtrus.org/2012-NPCR-SEER/CBTRUS_Report_2004-2008_3-23-2012.pdf.
- Yen KE, Bittinger MA, Su SM, Fantin VR. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene. 2010;29(49):6409–6417.
- Popovici-Muller J, et al. Discovery of the first potent inhibitors of mutant IDH1 that lower tumor 2-HG in vivo. ACS Med Chem Lett. 2012;3(10):850–855.View this article via: CrossRef Google Scholar
- Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744.
- Reitman ZJ, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A. 2011;108(8):3270–3275.
- Ward PS, et al. Identification of additional IDH mutations associated with oncometabolite R(-)-2-hydroxyglutarate production. Oncogene. 2012;31(19):2491–2498.
- Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30.
- Chowdhury R, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–469.
- Losman JA, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339(6127):1621–1625.
- Williams SC, Karajannis MA, Chiriboga L, Golfinos JG, von Deimling A, Zagzag D. R132H-mutation of isocitrate dehydrogenase-1 is not sufficient for HIF-1 alpha upregulation in adult glioma. Acta Neuropathologica. 2011;121(2):279–281.
- Bleeker FE, et al. The prognostic IDH1 (R132) mutation is associated with reduced NADP(+)-dependent IDH activity in glioblastoma. Acta Neuropathologica. 2011;119(4):487–494.
- Li S, et al. Overexpression of isocitrate dehydrogenase mutant proteins renders glioma cells more sensitive to radiation. Neuro Oncol. 2013;15(1):57–68.
- Beckonert O, et al. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc. 2010;5(6):1019–1032.
- Bal D, Gryff-Keller A. H-1 and C-13 NMR study of 2-hydroxyglutaric acid and its lactone. Magn Reson Chem. 2002;40(8):533–536.View this article via: CrossRef Google Scholar
- Pope WB, et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol. 2012;107(1):197–205.
- Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11(6):266–272.
- Dowling C, et al. Preoperative proton MR spectroscopic imaging of brain tumors: Correlation with histopathologic analysis of resection specimens. AJNR Am J Neuroradiol. 2001;22(4):604–612.View this article via: PubMed Google Scholar
- Howe FA, et al. Metabolic profiles of human brain tumors using quantitative in vivo H-1 magnetic resonance spectroscopy. Magn Reson Chem. 2003;49(2):223–232.
- Wright AJ, Fellows GA, Griffiths JR, Wilson M, Bell BA, Howe FA. Ex-vivo HRMAS of adult brain tumours: metabolite quantification and assignment of tumour biomarkers. Mol Cancer. 2010;9:66.
- Andronesi OC, et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4(116):116ra4.
- Choi C, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in subjects with IDH-mutated gliomas. Nat Med. 2012;18(4):624–629.
- Ernst RR. Two-dimensional spectroscopy. Chimia. 1975;29:179–183.
- Jeener J. Ampere International Summer School, Basko Polje, Yugoslavia, 1971. In: Golman M, Porneuf M, eds. NMR and More. Les Ulis, France: Les Editions de Physique; 1994.
- Rothman DL, Petroff OAC, Behar KL, Mattson RH. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proc Natl Acad Sci U S A. 1993;90(12):5662–5666.
- Wilman AH, Allen PS. Yield enhancement of a double-quantum filter sequence designed for the edited detection of GABA. J Magn Reson B. 1995;109(2):169–174.
- Shen J, Rothman DL, Brown P. In vivo GABA editing using a novel doubly selective multiple quantum filter. Magn Reson Med. 2002;47(3):447–454.
- Dreher W, Leibfritz D. Detection of homonuclear decoupled in vivo proton NMR spectra using constant time chemical shift encoding: CT-PRESS. Magn Reson Imaging. 1999;17(1):141–150.
- Schulte RF, Lange T, Beck J, Meier D, Boesiger P. Improved two-dimensional J-resolved spectroscopy. NMR Biomed. 2006;19(2):264–270.
- Thomas MA, et al. Localized two-dimensional shift correlated MR spectroscopy of human brain. Magn Reson Med. 2001;46(1):58–67.
- Andronesi OC, Ramadan S, Mountford CE, Sorensen AG. Low-power adiabatic sequences for in-vivo localized two-dimensional chemical shift correlated MR spectroscopy. Magn Reson Med. 2010;64(6):1542–1556.
- Bottomley PA. Spatial localization in Nmr-spectroscopy in vivo. Ann N Y Acad Sci. 1987;508:333–348.
- Near J, Simpson R, Cowen P, Jezzard P. Efficient γ-aminobutyric acid editing at 3T without macromolecule contamination: MEGA-SPECIAL. NMR Biomed. 2006;19(2):264–270.
- Zhu H, Edden RA, Ouwerkerk R, Barker PB. High resolution spectroscopic imaging of GABA at 3 Tesla. Magn Reson Med. 2011;65(3):603–609.
- Andronesi OC, Ramadan S, Ratai EM, Jennings D, Mountford CE, Sorensen AG. Spectroscopic imaging with improved gradient modulated constant adiabaticity pulses on high-field clinical scanners. J Magn Reson. 2010;203(2):283–293.
- Garwood M, DelaBarre L. The return of the frequency sweep: Designing adiabatic pulses for contemporary NMR. J Magn Reson. 2001;153(2):155–177.
- Marjanska M, et al. Brain dynamic neurochemical changes in dystonic patients: A magnetic resonance spectroscopy study. Mov Disord. 2013;28(2):201–209.
- Hess AT, Tisdall MD, Andronesi OC, Meintjes EM, van der Kouwe AJW. Real-time motion and B0 corrected single voxel spectroscopy using volumetric navigators. Magn Reson Med. 2011;66(2):314–323.
- Hess AT, Andronesi OC, Tisdall MD, van der Kouwe AJW, Sorensen AG, Meintjes E. Real-time Motion and B0 correction for LASER MRSI using EPI volumetric navigators. NMR Biomed. 2012;25(2):347–358.
- Maudsley AA, et al. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 2006;19(4):492–503.
- Xu V, et al. MR spectroscopy in diagnosis and neurological decision-making. Semin Neurol. 2008;28(4):407–422.
- Ernst RR, Bodenhausen G, Wokaun A. Principles of Nuclear Magnetic Resonance in One and Two Dimensions. Oxford, United Kingdom: Clarendon Press; 1987.
- Cheng LL, et al. Quantitative neuropathology by high resolution magic angle spinning proton magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 1997;94(12):6408–6413.
- Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat Rev Cancer. 2004;4(7):551–561.
- Andronesi OC, Mintzopoulos D, Struppe J, Black PM, Tzika AA. Solid-state NMR adiabatic TOBSY sequences provide enhanced sensitivity for multidimensional high-resolution magic-angle-spinning H-1 MR spectroscopy. J Magn Reson. 2008;193(2):251–258.
- Elkhaled A, et al. Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4(116):116ra5.
- Kalinina J, et al. Detection of “oncometabolite” 2-hydroxyglutarate by magnetic resonance analysis as a biomarker of IDH1/2 mutations in glioma. J Mol Med (Berl). 2012;90(10):1161–1171.
- Metellus P, et al. IDH mutation status impact on in vivo hypoxia biomarkers expression: new insights from a clinical, nuclear imaging and immunohistochemical study in 33 glioma patients. J Neurooncol. 2011;105(3):591–600.
- Kunz M, et al. Hot spots in dynamic (FET)-F-18-PET delineate malignant tumor parts within suspected WHO grade II gliomas. Neuro Oncol. 2011;13(3):307–316.
- Dunn GP, Andronesi OC, Cahill DP. From genomics to the clinic: biological and translational insights of mutant IDH1/2 in glioma. Neurosurg Focus. 2013;34(2):E2.View this article via: PubMed Google Scholar
- Capper D, Zentgraf H, Balss J, Hartmann C, von Deimling A. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol. 2009;118(5):599–601.
- Fathi AT, et al. Prospective serial evaluation of 2-hydroxyglutarate, during treatment of newly diagnosed acute myeloid leukemia, to assess disease activity and therapeutic response. Blood. 2012;120(23):4649–4652.
- Nakamizo S, et al. GC/MS-based metabolomic analysis of cerebrospinal fluid (CSF) from glioma patients. J Neurooncol. 2013;113(1):65–74.
-
Version history
- Version 1 (September 3, 2013): No description
Article tools
-
View PDF

- Download citation information
- Send a comment
- Terms of use
- Standard abbreviations
- Need help? Email the journal
Review Series
Cancer Metabolism
-
Oncometabolites: linking altered metabolism with cancer
Ming Yang et al. -
Detection of oncogenic IDH1 mutations using magnetic resonance spectroscopy of 2-hydroxyglutarate
Ovidiu C. Andronesi et al. -
HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations
Gregg L. Semenza -
Potential applications for biguanides in oncology
Michael Pollak -
Isocitrate dehydrogenase mutations in leukemia
Anna Sophia McKenney et al. -
Glutamine and cancer: cell biology, physiology, and clinical opportunities
Christopher T. Hensley et al. -
Targeting lactate metabolism for cancer therapeutics
Joanne R. Doherty et al. -
Exploiting tumor metabolism: challenges for clinical translation
Matthew G. Vander Heiden
Metrics
Go to



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)


