Advertisement
Review Free access | 10.1172/JCI43085
Glaucoma: genes, phenotypes, and new directions for therapy
Bao Jian Fan and Janey L. Wiggs
Massachusetts Eye and Ear Infirmary, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Janey L. Wiggs, Massachusetts Eye and Ear Infirmary, Harvard Medical School, 243 Charles St., Boston, Massachusetts, USA. Phone: 617.573.6440; Fax: 617.573.6439; E-mail: janey_wiggs@meei.harvard.edu.
Find articles by Fan, B. in: PubMed | Google Scholar
Massachusetts Eye and Ear Infirmary, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Janey L. Wiggs, Massachusetts Eye and Ear Infirmary, Harvard Medical School, 243 Charles St., Boston, Massachusetts, USA. Phone: 617.573.6440; Fax: 617.573.6439; E-mail: janey_wiggs@meei.harvard.edu.
Find articles by Wiggs, J. in: PubMed | Google Scholar
Published September 1, 2010 - More info
J Clin Invest. 2010;120(9):3064–3072. https://doi.org/10.1172/JCI43085.
© 2010 The American Society for Clinical Investigation
-
Abstract
Glaucoma, a leading cause of blindness worldwide, is characterized by progressive optic nerve damage, usually associated with intraocular pressure. Although the clinical progression of the disease is well defined, the molecular events responsible for glaucoma are currently poorly understood and current therapeutic strategies are not curative. This review summarizes the human genetics and genomic approaches that have shed light on the complex inheritance of glaucoma genes and the potential for gene-based and cellular therapies that this research makes possible.
-
Introduction
Glaucoma is a term used to describe a group of disorders that have in common a distinct type of optic nerve damage, causing irreversible defects in the visual field. The optic neuropathy is progressive and, if left untreated, results in absolute blindness. Glaucoma is a leading cause of blindness worldwide, affecting 2% of individuals of European descent and up to 10% of individuals of sub-Saharan African descent over 50 years of age (1). The disease process is insidious, and since central vision is usually not lost until the disease is advanced, a significant proportion of individuals remain either undiagnosed or undertreated. The most common forms of glaucoma are age-related, beginning in midlife and progressing slowly but relentlessly (2). If detected early enough, disease progression can be slowed with drug and/or surgical treatment, underscoring the importance of identifying the disease in its earliest stages (2).
Elevated intraocular pressure (IOP) is a major risk factor for glaucoma. Fluid formed by the ciliary body (aqueous humor) is removed by the trabecular outflow pathways, which includes the trabecular meshwork (made up by the uveal and corneoscleral meshworks), the juxtacanalicular connective tissue, the endothelial lining of Schlemm’s canal, and the collecting channels and the aqueous veins (2). The trabecular meshwork outflow pathways provides resistance to aqueous fluid outflow (3) and normally functions to maintain a minimum level of eye pressure that keeps the eye in the round shape necessary for its refractive properties. The IOP is dependent on the rate of fluid removal, which under normal conditions matches the rate of formation. In most patients with glaucoma, the rate of fluid removal declines so that it no longer keeps pace with the rate of formation, and the IOP increases. Some fluid can exit the eye through an alternative pathway (uveoscleral pathway); however, this is not sufficient to maintain homeostasis (Figure 1).
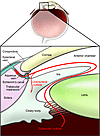 Figure 1
Figure 1Schematic diagram of eye structures involved in aqueous humor dynamics. Aqueous humor is produced by the ciliary body and passes from the posterior chamber through the pupil into the anterior chamber into the trabecular meshwork, Schlemm’s canal, and finally into the episcleral venous system. The alternative path is the “uveoscleral outflow” that drains fluid through the ciliary muscle into the supraciliary and suprachoroidal spaces and then out of the eye through the sclera. Approximately 80% of aqueous humor is removed by the conventional trabecular meshwork pathway, and the remainder is removed by the uveoscleral pathway. Elevated IOP in glaucoma is caused by alterations to the conventional trabecular meshwork pathway. The figure adapted from Berwick Eye and Surgicentre ( www.berwickeye.com.au/glaucoma.htm).
The molecular events responsible for a decrease in aqueous outflow in most patients with glaucoma are not known. Current treatment for glaucoma is directed toward lowering the IOP by reducing the amount of fluid made by the ciliary body, increasing the flow of fluid through the trabecular meshwork or uveoscleral pathways, and/or by surgically creating an alternative path for fluid egress. None of these approaches are curative, and over time multiple treatments are generally required to limit disease progression (4). Loss of visual function in glaucoma is caused by apoptosis of retinal ganglion cells, leading to a loss of ganglion cell axons and degeneration of the optic nerve. Retinal ganglion cell loss results in permanent visual field defects that progressively enlarge and can eventually cause total blindness. Ganglion cell loss is usually, but not always, associated with elevated IOP (5). Progressive ganglion cell loss without increased IOP is called normal tension glaucoma (NTG) (6).
The molecular events responsible for glaucoma are currently poorly understood, complicating the design of therapies based on the underlying disease mechanisms. Most forms of glaucoma are inherited, either as common complex traits or as mendelian autosomal dominant or recessive traits (early onset). This review summarizes the glaucoma genes and phenotypes identified using genetic and genomic approaches and the potential for gene-based and cellular therapies granted by this emerging science.
-
Genes identified using linkage approaches and multiplex families
Large multiplex families suitable for linkage analysis are typically affected by early-onset forms of glaucoma inherited as mendelian dominant or recessive traits. Most pedigrees affected by adult-onset forms of glaucoma do not have the sufficient size and structure required for linkage studies, because the older age of onset typically limits these pedigrees to one and possibly two affected generations (7). To date, at least 29 genetic loci for various forms of glaucoma have been defined by linkage studies (8, 9), and 12 causative genes have been identified from these loci (Table 1). Genes and loci for different phenotypic forms of glaucoma are summarized below.
Congenital glaucoma. The onset of congenital glaucoma is typically before 3 years of age and is characterized by very high IOP caused by abnormal development of the trabecular outflow pathways (10). This form of glaucoma is typically inherited as an autosomal recessive trait and is more common in countries and cultures that practice consanguinity. Linkage studies have identified four loci, GLC3A (2p22-p21) (11), GLC3B (1p36.2-p36.1) (12), GLC3C (14q24.3) (13), and GLC3D (14q24) (14), and two genes, CYP1B1, coding for cytochrome P450, family 1, subfamily B, polypeptide 1 at GLC3A (15), and more recently, LTBP2, coding for latent transforming growth factor-β–binding protein 2 at GLC3D (16, 17). Although known to cause congenital glaucoma for over a decade, the role of cytochrome P450 1B1 in glaucoma pathogenesis and development of the trabecular meshwork is not yet known but is hypothesized to participate in the metabolism of endogenous molecules that are necessary for ocular development (18, 19). Latent transforming growth factor-β–binding protein 2 is an ECM protein with homology to fibrillins and may participate in cell adhesion and may have a structural role in the development of the ocular outflow pathways (18, 19).
Anterior segment dysgenesis syndromes. The anterior segment of the eye includes all the structures in front of the ocular lens (iris, trabecular meshwork, anterior chamber, aqueous humor, and cornea). A majority of these structures arise from the cranial neural crest, and mutations in genes controlling ocular developmental processes result in abnormal formation of the trabecular meshwork and other structures necessary for fluid removal, causing an early onset (typically between 10 to 20 years of age) of very high IOP (20). These developmental disorders are usually inherited as autosomal dominant traits and may be associated with systemic abnormalities (20). For example, in addition to ocular dysgenesis, patients affected by Axenfeld-Rieger syndrome have abnormal teeth and hearing loss (21). Several loci and two genes coding for transcription factors (paired-like homeodomain transcription factor 2 [PITX2] at RIEG1, ref. 22, and forkhead box C1 [FOXC1] at RIEG3, ref. 23) have been associated with this syndrome. Other genes causing developmental glaucoma are PAX6 (Aniridia) (24) and LMX1B (nail-patella syndrome and anterior segment dysgenesis) (25, 26). Interestingly, the developmental glaucoma syndromes frequently exhibit variable expressivity, with glaucoma only occurring in about 50% of cases overall (27). The cause of the variable expressivity is not known; however, it may be secondary to modifier genes or stochastic effects (28).
Nanophthalmos. Nanophthalmos is a developmental disorder that causes the eye to be smaller than normal (29). The reduced size of the eye crowds the intraocular structures, which increases the risk of a form of glaucoma called angle-closure glaucoma (29). Three loci (NNO1–NNO3) and one gene (membrane frizzled-related protein [MFRP] at NNO2) have been identified for this condition (30–32). The function of the MFRP protein is not completely understood; however, it appears to be necessary for retinal development, particularly the photoreceptor layer of the retina (33). When the retinal photoreceptors do not form normally, the eye is smaller, shorter, and predisposed to angle-closure glaucoma (33).
Pigment dispersion syndrome and pigmentary glaucoma. Pigment dispersion syndrome is an ocular condition characterized by shedding of pigment granules from the iris, which become distributed throughout the eye, including the trabecular outflow pathways (34). The presence of pigment in the outflow pathways causes an increase in IOP and glaucoma in approximately 50% of individuals affected with the condition (34). The condition can be inherited as an autosomal dominant trait, and one loci (GPDS1) has been identified; however, the causative gene has not yet been discovered (35). The DBA/2J mouse spontaneously develops glaucoma that is clinically similar to human pigmentary glaucoma; however, the genes responsible for the mouse phenotype do not cause the disease in humans (36, 37).
Juvenile open-angle glaucoma. The term open-angle glaucoma refers to forms of glaucoma occurring in eyes with normal anatomic structures. Usually, open-angle glaucoma develops in adults after 50 years of age (called primary open-angle glaucoma [POAG]), but rarely this type of glaucoma can develop in a child or young adult before 35 years of age. This early-onset form of POAG, called JOAG, is inherited as an autosomal dominant trait (38) and is characterized by extremely high IOP, usually requiring surgical treatment (39, 40). A number of genetic loci have been identified for this condition, GLC1A (1q24.3-q25.2) (41), GLC1J (9q22) (42), GLC1K (20p12) (42), GLC1M (5q22.1-q32) (43–45), and GLC1N (15q22-q24) (46), and one gene, MYOC, coding for Myocilin, has been identified from the GLC1A locus (47, 48). Myocilin is an extracellular protein of unknown ocular function. Missense changes in the protein account for 10% of the cases of JOAG and also 3%–5% of the cases of adult-onset POAG (49, 50). A loss of protein function does not result in glaucoma (51, 52), and the underlying genetic mechanism is likely to be a gain-of-function or dominant-negative effect (53). The disease-associated missense changes reduce the solubility of the protein, causing it to aggregate in the endoplasmic reticulum and preventing its secretion to the ECM (54). Since an absence of protein does not cause disease, it remains unknown why the aggregation of myocilin intracellularly results in elevated IOP; however, recent work suggests that accumulation of myocilin aggregates intracellularly may sensitize trabecular meshwork cells to apoptosis (55).
Adult-onset POAG. Many patients with adult-onset POAG have affected family members, making model-independent (genetic linkage studies that do not assume a specific inheritance model and are usually sibpair based) and model-dependent linkage approaches (linkage studies that require an inheritance model, i.e., autosomal dominant or autosomal recessive, to be defined) to gene identification possible. Model-dependent linkage analyses using multiplex POAG pedigrees have yielded a number of potential POAG loci (GLC1A–GLC1H and GLC1L; Table 1) but few genes. Optineurin (OPTN) at GLC1E (10p15-p14) (56), although initially described as a POAG-causative gene, is primarily responsible for rare cases of familial NTG, a type of open-angle glaucoma in which the optic nerve deteriorates despite normal IOP (57, 58). Optineurin may influence ganglion cell apoptosis directly through rab8 signaling (59, 60). WD repeat domain 36 (WDR36) at GLC1G (5q21.3-q22.1) appears to contribute to POAG severity in some cases but is not necessary or sufficient for disease development (61, 62). A zebrafish homolog of WDR36 stimulates apoptosis mediated by p53, suggesting a role for WDR36 in retinal ganglion cell susceptibility to apoptotic cell death (63). Genome-wide scans using nonparametric (model-free) linkage methods have also been completed for POAG pedigrees of European descent (64), Caribbean pedigrees of African descent in Barbados (65), and African-American pedigrees with diabetes and elevated IOP (66). These scans, using between 373 and 445 microsatellite repeat markers, identified 10 genomic regions that may harbor POAG susceptibility genes (2p14, 2q33-34, 10-12-p13, 14q11-q22, 17p13, 17q25, 19q12-q14). A follow-up study of the scan performed on pedigrees of European descent, using ordered subset analysis with the mean family age of onset as a covariate, identified a subset of 15 families with significant linkage to 15q11-q13, designated GLC1I (67). Recent studies suggest that the neurotrophin 4 (NTF4) gene located in the 19p13.3 region (GLC1O) may contribute to POAG in a German population (68). Alterations in neurotrophin 4 are predicted to have decreased affinity with its specific receptor, tyrosine kinase receptor B (TrkB), leading to decreased activation of TrkB. These findings suggest that loss of neurotrophic function may be one of the general mechanisms involved in the pathogenesis of glaucoma and that agents activating TrkB could be considered as neuroprotective targets.
-
Case-control association studies: POAG and NTG
It is likely that multiple genes and/or environmental factors contribute to the complex inheritance of adult-onset glaucoma and that different sets of genetic and environmental risk factors contribute to regulation of IOP and ganglion cell apoptosis. A number of candidate genes have been evaluated for association with POAG and NTG; however, the strength of association is variable among these studies, and most have not been replicated in other studies or populations (Table 2). A gene-environment interaction has been observed between nitric oxide synthetase 3 (NOS3) SNPs and female sex and postmenopausal hormone use in the women in relation to POAG (69). Other gene-environment and gene-gene interactions are likely to contribute to POAG.
-
Genome-wide association studies
Exfoliation syndrome and glaucoma. Of the adult-onset forms of glaucoma, a sufficiently powered genome-wide association study has only been completed for exfoliation glaucoma. Exfoliation syndrome, characterized by the deposition of microfibrillar material throughout the eye, is a significant risk factor for glaucoma, with over 50% of affected individuals developing IOP and optic nerve degeneration by 70 years of age (70). Exfoliation syndrome is inherited as a complex trait, and a genome-wide association study using cases and controls from Iceland initially identified lysyl oxidase-like 1 (LOXL1) as a major genetic risk factor for the condition (71), with a population-attributable risk of more than 99%. Subsequently, this association has been replicated in populations throughout the world (72–76). Importantly, although the risk allele (G153D) is present in the majority of cases, it is also highly prevalent in controls, indicating that LOXL1 is necessary but not sufficient for the disease and that other genetic and/or environmental factors are required for disease development (77). LOXL1 is a member of the lysyl oxidase family of proteins that catalyze the polymerization of tropoelastin to form the mature elastin polymer (78). Elastin fibers are a major component of many structures in the eye, such as the ECM of the trabecular meshwork and the lamina cribrosa of the optic nerve (79, 80). LOXL1 is also involved in elastin homeostasis and renewal and participates in spatially organizing elastogenesis at sites of elastin deposition (81). The role of the G153D risk allele in disease development is not yet clear, and recent studies suggest that the risk allele in the black South African population is reversed compared with other populations, arguing that neither the Glycine allele or the Aspartate allele is biologically causative (82). Interestingly, the LOXL1-null mouse has some features of the human disease, including lens and iris abnormalities, but does not exhibit increased IOP or optic nerve damage (unpublished observations).
POAG. Genome-wide association studies in Japanese populations (83) and Caribbean populations of African descent (84) have identified (SNPs) with possible POAG association, but these have not revealed specific gene associations. Future genome-wide association studies with sufficient power to detect genes of moderate effect will provide important information about the genes contributing to this complex condition (unpublished observations).
-
Pathophysiology associated with current glaucoma genes
Most of the current glaucoma genes cause early-onset forms of glaucoma characterized by severely elevated IOP. Most of these genes (CYP1B1, LTPB2, PITX2, FOXC1, PAX6, and LMX1B) modulate ocular developmental processes, and gene mutations lead to abnormal development of the anterior segment structures necessary for aqueous humor removal (20). The MYOC gene, coding for myocilin, is the only gene currently known to cause elevated IOP with normal development of the ocular anterior segment (40). It is expected that certain glaucoma-predisposing genes will specifically influence ganglion cell death in the setting of elevated IOP or even normal IOP. Ganglion cell apoptosis is associated with a mutant form of optineurin (56, 57), and WDR36 variants may increase the degree of ganglion cell loss (62). Gene-specific pathophysiology has not yet been established for other glaucoma-causing genes.
Glaucoma mouse models of several of the human glaucoma genes have also helped define the pathological processes responsible for disease (85). CYP1B1-null mice exhibit abnormal development of the ocular anterior segment and aqueous outflow structures (86). Interestingly, tyrosinase modifies the severity of the developmental abnormalities in mice but not in humans (87). FOXC1-deficient mice have ocular defects that are similar to those found in humans with FOXC1 mutations (88). MYOC-null mice do not have elevated IOP (52), while transgenic mice expressing a human missense mutation do develop glaucoma (89). The role of OPTN in ganglion cell loss is supported by studies showing that overexpression of the optineurin E50K mutation in mice causes loss of ganglion cells without elevation of IOP (90). In addition to confirming the biological significance of mutant forms of these genes, future studies of these mouse models will help evaluate novel therapeutic approaches for the disease (85).
-
Quantitative traits
Many heritable ocular traits are quantitative, and many of these are risk factors for glaucoma, including central corneal thickness, IOP, and anatomic features of the optic nerve. The heritability estimate, defined as the proportion of variation between individuals in a population that is influenced by genetic factors, is 0.35 for IOP, 0.55 for optic cup diameter, 0.57 for optic disc diameter, 0.58 for vertical cup-to-disc ratio, 0.39 for neuroretinal rim area, 0.48 for retinal nerve fiber layer thickness, and 0.68 for central corneal thickness (91–94).
A variety of studies have been performed to identify genetic determinants of ocular quantitative traits, including genome-wide linkage analyses for IOP, genome-wide association studies for optic nerve parameters, and gene-association studies for central corneal thickness. A number of genomic regions have been identified as IOP linkage regions (95), and several genes have been identified as possible candidates for influencing central corneal thickness (96), including common polymorphisms in the type I collagen genes, COL1A1 and COL1A2. Two of the IOP regions on chromosomes 2 and 19 overlap with regions identified as potential loci for blood pressure, suggesting factors that may participate in regulation of both IOP and blood pressure (95). Recently a genome-wide association study has identified a SNP located near the ATOH7 gene as a significant determinant of optic nerve size (97).
-
Biomarkers for glaucoma
Individuals predisposed to glaucoma should be identified at the earliest possible stages of the disease, so that treatment can be initiated before the optic nerve is irreversibly damaged. Although current therapy is not curative, for many patients lowering the IOP using currently available pharmaceutical and/or surgical approaches can slow the progression of the disease and limit the overall loss of vision. Thus, biomarkers might be invaluable tools to identify individuals at risk for disease and, depending on the approach, could serve to measure the outcomes of therapies. The development of clinically useful biomarkers is an area of active investigation and includes genetic screening tests, proteomic markers, and analyses of serum antibodies to retina and optic nerve proteins.
Genetic screening tests. Currently, genetic testing can identify individuals at risk for early-onset forms of glaucoma in approximately 30% of cases (98). Useful gene-based screening tests for adult-onset POAG are not yet available (99); however, ongoing studies using large case/control cohorts for genome-wide association, such as those discussed above, may reveal important genetic determinants that could lead to clinically relevant screening tests. Although a major gene (LOXL1) has been associated with adult-onset exfoliation glaucoma, use of the gene variants for genetic screening remains challenging. The high prevalence of the LOXL1 risk genotype in control populations confers an extremely low specificity, making it almost useless as a predictive screening test (77). The specificity of genetic testing is increased with a combination of the LOXL1 risk genotype and a variant in the 5' promoter region; however, the positive predictive value remains low, as the specific 5' variant is rare (unpublished observations).
Proteomic biomarkers. Proteomic approaches using serum, tears, and aqueous humor could identify useful glaucoma biomarkers (100). Analysis of serum antibodies to optic nerve or retinal proteins may detect glaucoma-specific autoantibodies that could be indicative of ganglion cell damage (101). However, the small amount of tissue affected by the disease is a limitation of using proteomics to identify a glaucoma biomarker with the specificity and sensitivity required for clinical use. Proteomic approaches may also be comprised by the confounding effects of concurrent medical and/or surgical treatments, as these therapies can also influence the protein composition of affected tissues (102).
-
Experimental therapeutic approaches
In glaucoma, loss of vision is caused by degeneration of retinal ganglion cells. Current treatment is directed toward lowering IOP, which in most patients is related to the rate and extent of ganglion cell loss. Although IOP can be controlled pharmacologically and surgically, no treatment is currently available to prevent and restore retinal and optic nerve function. In addition to lowering IOP, neuroprotective strategies could effectively preserve vision in glaucoma. A variety of experimental approaches to promote retinal ganglion cell survival are underway, including intraocular delivery of neuroprotective molecules, viral-mediated gene transfer, and stem cell therapies (Figure 2).
 Figure 2
Figure 2Schematic overview of experimental therapeutic approaches to glaucoma, including intraocular delivery of neuroprotective molecules, viral-mediated gene transfer, and stem cell therapies.
Intraocular delivery of neuroprotective molecules. Neurotrophic agents are known to promote survival of retinal ganglion cells (103). In animal models, intravitreal injections of brain-derived neurotrophic factor can rescue retinal ganglion cells after optic nerve injury (104), although the long-term effects of those treatments and overall risks are not known (105). Recent experiments suggest that combinations of growth factors, such as the combination of brain-derived neurotrophic factor and LINGO-1 fusion protein, can provide more complete rescue of ganglion cells for longer periods of time (106). In addition, delivery of ciliary-derived neurotrophic factor, using viral transfer and injections of nerve growth factor in the episcleral veins, has been shown to be neuroprotective in a rat model of glaucoma (107, 108). Although current investigations are limited to animal models, these studies suggest that future clinical applications in human glaucoma patients could prevent ganglion cell damage and restore ganglion cell function.
Gene replacement therapies. Gene replacement therapies may specifically target the underlying molecular events responsible for the disease and, depending on the choice of viral vector, may have long-lasting local effects. The eye is easily accessible, making injectable viral-transfer methods an attractive therapeutic option. Indeed, the success of a viral-based gene delivery approach has been demonstrated in an ocular disease, Leber’s congenital amaurosis, caused by mutations in a retinal protein, RPE65 (109). As genetic studies continue to elucidate the underlying molecular events responsible for glaucoma, the use of gene-based therapies for pathology of the trabecular meshwork and of the retinal ganglions cells are gaining support (110). Experimental gene-based therapies targeting the trabecular meshwork include the overexpression of recombinant matrix metalloproteinase I (MMP1) to counteract ECM deposition in patients affected by steroid-induced glaucoma (111). Lentiviral vectors have been used to deliver COX-2, a regulator of prostaglandin formation, to increase uveal scleral outflow of fluid (112). Self-complementary AAV has been shown to provide safe and long-term gene transfer to the trabecular meshwork in living rats and monkeys, making stable expression with minimal risks possible (113). Finally, intravitreal injection of viral constructs carrying neurotrophic factors have also successfully prevented ganglion cell loss in animal models of glaucoma (107), suggesting that viral gene-based therapy may be a viable approach to protect ganglion cells in glaucoma patients.
Stem cell–based therapy. Stem cells may be useful therapeutic agents for neurodegenerative diseases, because they congregate at the sites of injury and may provide chronic neuroprotection after a single treatment (114). Stem cell transplantation can supply neurotrophic factors and also modulate other components of the CNS environment to facilitate endogenous repair. Considerable progress in the understanding of the pathways involved in directed axon growth and the manipulation of stem and progenitor cells toward a RGC fate (115) suggest that stem cell therapies using mesenchymal stem cells and neural stem cells for glaucoma could be possible (116). Intravitreal injection of systemic bone marrow–derived mesenchymal stem cells in a rat glaucoma model resulted in an increase in overall RGC axon survival (117), and injection of neural stem cells (oligodendrocyte precursor cells) was also neuroprotective (118). In addition, at least one study suggests that stem cell transplantation may also improve fluid removal through the trabecular meshwork (119). Though many challenges remain, including identifying sources for suitable stem cells and determining the safety of therapeutic stem cell use, these and other recent studies suggest that stem cell transplantation could be a promising therapeutic approach for glaucoma.
-
Future directions
Novel cell-based and viral-gene-transfer therapies promise to bring new hope for visual preservation, but they may also bring significant risk. It will be important to carefully select patients who will benefit from these therapies. The ocular hypertensive treatment study, a clinical trial designed to evaluate the impact of topical IOP-lowering therapy on the onset of visual field loss and/or optic nerve damage in patients with elevated IOP, showed that only 10%–15% of patients with elevated IOP actually develop optic nerve disease (120, 121), and treatment should be targeted to those at increased risk for visual loss, which includes those individuals with older age, larger vertical or horizontal cup-disc ratio, higher IOP, greater visual field loss, and thinner central corneal measurement. Sufficiently sensitive and specific screening tools (genetic and/or proteomic biomarkers) will be needed to effectively identify individuals who will benefit from therapy. It is also important that treatment is initiated when it will have the most benefit. Technologies that will make it possible to monitor “sick” ganglion cells at stages when they can still be rescued need to be developed, and recent advances in retinal imaging, such as spectral domain optical coherence tomography, are promising (122). This technique creates a 3-dimensional image of the optic nerve, with sufficient resolution that structural changes in the optic nerve caused by glaucoma could be detected at early stages when therapeutic options could have maximal benefit. Real-time in vivo imaging of retinal cell apoptosis using intravitreal fluorescence-labeled annexin 5, followed by microscopy, suggests that methods to identify ganglion cells entering apoptosis in glaucoma patients may be possible in the future (123). These technologies will also make it possible to determine the efficacy of therapy. In the future, we can expect that a patient’s risk for glaucoma will be established using genetic and other screening tests, an assessment of ganglion cell disease will be made by novel imaging technologies, and appropriate therapy will be initiated to restore ganglion cell health. Extension of the genetic studies summarized here will allow early disease detection and timely therapy targeted to molecular disease mechanisms, granting glaucoma patients the best chance to maintain useful sight.
-
Acknowledgments
This work was supported in part by NIH grants R01EY009847, R01EY015872, R21EY019161, and P30EY014104 and by Research to Prevent Blindness and the Massachusetts Eye Research Fund.
Address correspondence to: Janey L. Wiggs, Massachusetts Eye and Ear Infirmary, Harvard Medical School, 243 Charles St., Boston, Massachusetts, USA. Phone: 617.573.6440; Fax: 617.573.6439; E-mail: janey_wiggs@meei.harvard.edu.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2010;120(9):3064–3072. doi:10.1172/JCI43085
-
References
- Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90(3):262–267.
- Ophthalmology. 2009;116(4):710–716.
- Tamm ER. The trabecular meshwork outflow pathways: structural and functional aspects. Exp Eye Res. 2009;88(4):648–655.
- Mosaed S, Dustin L, Minckler DS. Comparative outcomes between newer and older surgeries for glaucoma. Trans Am Ophthalmol Soc. 2009;107:127–133.View this article via: PubMed Google Scholar
- Qu J, Wang D, Grosskreutz CL. Mechanisms of retinal ganglion cell injury and defense in glaucoma. Exp Eye Res. 2010;91(1):48–53.
- Cheng JW, Cai JP, Wei RL. Meta-analysis of medical intervention for normal tension glaucoma. Ophthalmology. 2009;116(7):1243–1249.
- Wiggs JL, Damji KF, Haines JL, Pericak-Vance MA, Allingham RR. The distinction between juvenile and adult-onset primary open-angle glaucoma. Am J Hum Genet. 1996;58(1):243–244.View this article via: PubMed Google Scholar
- Fan BJ, Wang DY, Lam DS, Pang CP. Gene mapping for primary open angle glaucoma. Clin Biochem. 2006;39(3):249–258.
- Wiggs JL. Genetic etiologies of glaucoma. Arch Ophthalmol. 2007;125(1):30–37.
- Sarfarazi M, Stoilov I. Molecular genetics of primary congenital glaucoma. Eye (Lond). 2000;14(pt 3B):422–428.View this article via: PubMed Google Scholar
- Sarfarazi M, et al. Assignment of a locus (GLC3A) for primary congenital glaucoma (Buphthalmos) to 2p21 and evidence for genetic heterogeneity. Genomics. 1995;30(2):171–177.
- Akarsu AN, et al. A second locus (GLC3B) for primary congenital glaucoma (Buphthalmos) maps to the 1p36 region. Hum Mol Genet. 1996;5(8):1199–1203.
- Stoilov IR, Sarfarazi M. The third genetic locus (GLC3C) for primary congenital glaucoma (PCG) maps to chromosome 14q24.3. Invest Ophthalmol Vis Sci. 2002;43:e3015.
- Firasat S, Riazuddin SA, Hejtmancik JF, Riazuddin S. Primary congenital glaucoma localizes to chromosome 14q24.2-24.3 in two consanguineous Pakistani families. Mol Vis. 2008;14:1659–1665.View this article via: PubMed Google Scholar
- Stoilov I, Akarsu AN, Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet. 1997;6(4):641–647.
- Ali M, et al. Null mutations in LTBP2 cause primary congenital glaucoma. Am J Hum Genet. 2009;84(5):664–671.
- Narooie-Nejad M, et al. Loss of function mutations in the gene encoding latent transforming growth factor beta binding protein 2, LTBP2, cause primary congenital glaucoma. Hum Mol Genet. 2009;18(20):3969–3977.
- Bejjani BA, et al. Mutations in CYP1B1, the gene for cytochrome P4501B1, are the predominant cause of primary congenital glaucoma in Saudi Arabia. Am J Hum Genet. 1998;62(2):325–333.
- Vasiliou V, Gonzalez FJ. Role of CYP1B1 in glaucoma. Annu Rev Pharmacol Toxicol. 2008;48:333–358.
- Gould DB, Smith RS, John SW. Anterior segment development relevant to glaucoma. Int J Dev Biol. 2004;48(8–9):1015–1029.
- Grosso S, et al. Familial Axenfeld-Rieger anomaly, cardiac malformations, and sensorineural hearing loss: a provisionally unique genetic syndrome? Am J Med Genet. 2002;111(2):182–186.
- Semina EV, et al. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14(4):392–399.View this article via: PubMed Google Scholar
- Mears AJ, et al. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am J Hum Genet. 1998;63(5):1316–1328.
- van Heyningen V, Williamson KA. PAX6 in sensory development. Hum Mol Genet. 2002;11(10):1161–1167.View this article via: PubMed Google Scholar
- Dreyer SD, et al. Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nat Genet. 1998;19(1):47–50.
- Mimiwati Z, et al. Nail-patella syndrome and its association with glaucoma: a review of eight families. Br J Ophthalmol. 2006;90(12):1505–1509.
- Saleem RA, Walter MA. The complexities of ocular genetics. Clin Genet. 2002;61(2):79–88.
- Hjalt TA, Semina EV. Current molecular understanding of Axenfeld-Rieger syndrome. Expert Rev Mol Med. 2005;7(25):1–17.View this article via: PubMed Google Scholar
- Yalvac IS, Satana B, Ozkan G, Eksioglu U, Duman S. Management of glaucoma in patients with nanophthalmos. Eye (Lond). 2008;22(6):838–843.View this article via: PubMed Google Scholar
- Othman MI, et al. Autosomal dominant nanophthalmos (NNO1) with high hyperopia and angle-closure glaucoma maps to chromosome 11. Am J Hum Genet. 1998;63(5):1411–1418.
- Sundin OH, et al. Extreme hyperopia is the result of null mutations in MFRP, which encodes a Frizzled-related protein. Proc Natl Acad Sci U S A. 2005;102(27):9553–9558.
- Li H, et al. Localization of a novel gene for congenital nonsyndromic simple microphthalmia to chromosome 2q11-14. Hum Genet. 2008;122(6):589–593.
- Won J, et al. Membrane frizzled-related protein is necessary for the normal development and maintenance of photoreceptor outer segments. Vis Neurosci. 2008;25(4):563–574.View this article via: PubMed Google Scholar
- Niyadurupola N, Broadway DC. Pigment dispersion syndrome and pigmentary glaucoma--a major review. Clin Experiment Ophthalmol. 2008;36(9):868–882.View this article via: PubMed Google Scholar
- Andersen JS, et al. A gene responsible for the pigment dispersion syndrome maps to chromosome 7q35-q36. Arch Ophthalmol. 1997;115(3):384–388.View this article via: PubMed Google Scholar
- Anderson MG, et al. Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice. Nat Genet. 2002;30(1):81–85.
- Lynch S, Yanagi G, DelBono E, Wiggs JL. DNA sequence variants in the tyrosinase-related protein 1 (TYRP1) gene are not associated with human pigmentary glaucoma. Mol Vis. 2002;8:127–129.View this article via: PubMed Google Scholar
- Wiggs JL, Damji KF, Haines JL, Pericak-Vance MA, Allingham RR. The distinction between juvenile and adult-onset primary open-angle glaucoma. Am J Hum Genet. 1996;58(1):243–244.View this article via: PubMed Google Scholar
- Johnson AT, et al. Clinical phenotype of juvenile-onset primary open-angle glaucoma linked to chromosome 1q. Ophthalmology. 1996;103(5):808–814.View this article via: PubMed Google Scholar
- Wiggs JL, Del Bono EA, Schuman JS, Hutchinson BT, Walton DS. Clinical features of five pedigrees genetically linked to the juvenile glaucoma locus on chromosome 1q21-q31. Ophthalmology. 1995;102(12):1782–1789.View this article via: PubMed Google Scholar
- Sheffield VC, et al. Genetic linkage of familial open angle glaucoma to chromosome 1q21-q31. Nat Genet. 1993;4(1):47–50.View this article via: PubMed Google Scholar
- Wiggs JL, et al. A genomewide scan identifies novel early-onset primary open-angle glaucoma loci on 9q22 and 20p12. Am J Hum Genet. 2004;74(6):1314–1320.View this article via: PubMed Google Scholar
- Wang DY, et al. Absence of myocilin and optineurin mutations in a large Philippine family with juvenile onset primary open angle glaucoma. Mol Vis. 2004;10:851–856.View this article via: PubMed Google Scholar
- Pang CP, et al. A genome-wide scan maps a novel juvenile-onset primary open angle glaucoma locus to chromosome 5q. Mol Vis. 2006;12:85–92.View this article via: PubMed Google Scholar
- Fan BJ, et al. Fine mapping of new glaucoma locus GLC1M and exclusion of neuregulin 2 as the causative gene. Mol Vis. 2007;13:779–784.View this article via: PubMed Google Scholar
- Wang DY, et al. A genome-wide scan maps a novel juvenile-onset primary open-angle glaucoma locus to 15q. Invest Ophthalmol Vis Sci. 2006;47(12):5315–5321.
- Stone EM, et al. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275(5300):668–670.
- Kubota R, et al. A novel myosin-like protein (myocilin) expressed in the connecting cilium of the photoreceptor: molecular cloning, tissue expression, and chromosomal mapping. Genomics. 1997;41(3):360–369.
- Wiggs JL, et al. Prevalence of mutations in TIGR/Myocilin in patients with adult and juvenile primary open-angle glaucoma. Am J Hum Genet. 1998;63(5):1549–1552.
- Fingert JH, et al. Analysis of myocilin mutations in 1703 glaucoma patients from five different populations. Hum Mol Genet. 1999;8(5):899–905.
- Wiggs JL, Vollrath D. Molecular and clinical evaluation of a patient hemizygous for TIGR/MYOC. Arch Ophthalmol. 2001;119(11):1674–1678.View this article via: PubMed Google Scholar
- Kim BS, et al. Targeted disruption of the myocilin gene (Myoc) suggests that human glaucoma-causing mutations are gain of function. Mol Cell Biol. 2001;21(22):7707–7713.
- Resch ZT, Fautsch MP. Glaucoma-associated myocilin: a better understanding but much more to learn. Exp Eye Res. 2009;88(4):704–712.
- Aroca-Aguilar JD, Martínez-Redondo F, Sánchez-Sánchez F, Coca-Prados M, Escribano J. Functional role of proteolytic processing of recombinant myocilin in self-aggregation. Invest Ophthalmol Vis Sci. 2010;51(1):72–78.
- Joe MK, Tomarev SI. Expression of myocilin mutants sensitizes cells to oxidative stress-induced apoptosis. Implication for glaucoma pathogenesis. Am J Pathol. 2010;176(6):2880–2890.
- Rezaie T, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295(5557):1077–1079.
- Hauser MA, et al. Distribution of optineurin sequence variations in an ethnically diverse population of low-tension glaucoma patients from the United States. J Glaucoma. 2006;15(5):358–363.
- Aung T, et al. Clinical features and course of patients with glaucoma with the E50K mutation in the optineurin gene. Invest Ophthalmol Vis Sci. 2005;46(8):2816–2822.
- Chalasani ML, Radha V, Gupta V, Agarwal N, Balasubramanian D, Swarup G. A glaucoma-associated mutant of optineurin selectively induces death of retinal ganglion cells which is inhibited by antioxidants. Invest Ophthalmol Vis Sci. 2007;48(4):1607–1614.
- Chibalina MV, Roberts RC, Arden SD, Kendrick-Jones J, Buss F. Rab8-optineurin-myosin VI: analysis of interactions and functions in the secretory pathway. Methods Enzymol. 2008;438:11–24.View this article via: PubMed Google Scholar
- Monemi S, et al. Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Hum Mol Genet. 2005;14(6):725–733.
- Hauser MA, et al. Distribution of WDR36 DNA sequence variants in patients with primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2006;47(6):2542–2546.View this article via: PubMed Google Scholar
- Skarie JM, Link BA. The primary open-angle glaucoma gene WDR36 functions in ribosomal RNA processing and interacts with the p53 stress-response pathway. Hum Mol Genet. 2008;17(16):2474–2485.
- Wiggs JL, et al. Genome-wide scan for adult onset primary open angle glaucoma. Hum Mol Genet. 2000;9(7):1109–1117.
- Nemesure B, et al. A genome-wide scan for primary open-angle glaucoma (POAG): the Barbados Family Study of Open-Angle Glaucoma. Hum Genet. 2003;112(5–6):600–609.View this article via: PubMed Google Scholar
- Rotimi CN, et al. Genomewide scan and fine mapping of quantitative trait loci for intraocular pressure on 5q and 14q in West Africans. Invest Ophthalmol Vis Sci. 2006;47(8):3262–3267.
- Allingham RR, et al. Early adult-onset POAG linked to 15q11-13 using ordered subset analysis. Invest Ophthalmol Vis Sci. 2005;46(6):2002–2005.
- Pasutto F, et al. Heterozygous NTF4 mutations impairing neurotrophin-4 signaling in patients with primary open-angle glaucoma. Am J Hum Genet. 2009;85(4):447–456.
- Kang JH, et al. Endothelial nitric oxide synthase gene variants and primary open-angle glaucoma: interactions with sex and postmenopausal hormone use. Invest Ophthalmol Vis Sci. 2010;51(2):971–979.
- Konstas AG, Mantziris DA, Stewart WC. Diurnal intraocular pressure in untreated exfoliation and primary open-angle glaucoma. Arch Ophthalmol. 1997;115(2):182–185.View this article via: PubMed Google Scholar
- Thorleifsson G, et al. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007;317(5843):1397–1400.
- Fan BJ, et al. DNA sequence variants in the LOXL1 gene are associated with pseudoexfoliation glaucoma in a U.S. clinic-based population with broad ethnic diversity. BMC Med Genet. 2008;9:5.
- Pasutto F, et al. Association of LOXL1 common sequence variants in German and Italian patients with pseudoexfoliation syndrome and pseudoexfoliation glaucoma. Invest Ophthalmol Vis Sci. 2008;49(4):1459–1463.
- Hewitt AW, et al. Ancestral LOXL1 variants are associated with pseudoexfoliation in Caucasian Australians but with markedly lower penetrance than in Nordic people. Hum Mol Genet. 2008;17(5):710–716.
- Lee KY, et al. Association of LOXL1 polymorphisms with pseudoexfoliation in the Chinese. Mol Vis. 2009;15:1120–1126.View this article via: PubMed Google Scholar
- Ozaki M, et al. Association of LOXL1 gene polymorphisms with pseudoexfoliation in the Japanese. Invest Ophthalmol Vis Sci. 2008;49(9):3976–3980.
- Wiggs JL. Association Between LOXL1 and pseudoexfoliation. Arch Ophthalmol. 2008;126(3):420–421.
- Liu X, et al. Elastic fiber homeostasis requires lysyl oxidase-like 1 protein. Nat Genet. 2004;36(2):178–18.
- Acott TS, Kelley MJ. Extracellular matrix in the trabecular meshwork. Exp Eye Res. 2008;86(4):543–561.
- Urban Z, Agapova O, Hucthagowder V, Yang P, Starcher BC, Hernandez MR. Population differences in elastin maturation in optic nerve head tissue and astrocytes. Invest Ophthalmol Vis Sci. 2007;48(7):3209–3215.
- Thomassin L, et al. The Pro-regions of lysyl oxidase and lysyl oxidase-like 1 are required for deposition onto elastic fibers. J Biol Chem. 2005;280(52):42848–42855.View this article via: PubMed Google Scholar
- Williams SE, et al. Major LOXL1 risk allele is reversed in exfoliation glaucoma in a black South African population. Mol Vis. 2010;16:705–712.View this article via: PubMed Google Scholar
- Nakano M, et al. Three susceptible loci associated with primary open-angle glaucoma identified by genome-wide association study in a Japanese population. Proc Natl Acad Sci U S A. 2009;106(31):12838–12842.View this article via: PubMed Google Scholar
- Jiao X, et al. Common variants on chromosome 2 and risk of primary open-angle glaucoma in the Afro-Caribbean population of Barbados. Proc Natl Acad Sci U S A. 2009;106(40):17105–17110.
- Lindsey JD, Weinreb RN. Elevated intraocular pressure and transgenic applications in the mouse. J Glaucoma. 2005;14(4):318–320.
- Libby RT, et al. Modification of ocular defects in mouse developmental glaucoma models by tyrosinase. Science. 2003;299(5612):1578–1581.
- Bidinost C, et al. Of mice and men: tyrosinase modification of congenital glaucoma in mice but not in humans. Invest Ophthalmol Vis Sci. 2006;47(4):1486–1490.View this article via: PubMed Google Scholar
- Smith RS, et al. Haploinsufficiency of the transcription factors FOXC1 and FOXC2 results in aberrant ocular development. Hum Mol Genet. 2000;9(7):1021–1032.
- Zhou Y, Grinchuk O, Tomarev SI. Transgenic mice expressing the Tyr437His mutant of human myocilin protein develop glaucoma. Invest Ophthalmol Vis Sci. 2008;49(5):1932–1939.
- Chi ZL, et al. Overexpression of optineurin E50K disrupts Rab8 interaction and leads to a progressive retinal degeneration in mice. Hum Mol Genet. 2010;19(13):2606–2615.
- van Koolwijk LM, et al. Genetic contributions to glaucoma: heritability of intraocular pressure, retinal nerve fiber layer thickness, and optic disc morphology. Invest Ophthalmol Vis Sci. 2007;48(8):3669–3676.View this article via: PubMed Google Scholar
- van Koolwijk LM, et al. Major genetic effects in glaucoma: commingling analysis of optic disc parameters in an older Australian population. Invest Ophthalmol Vis Sci. 2009;50(11):5275–5280.
- Klein BE, Klein R, Lee KE. Heritability of risk factors for primary open-angle glaucoma: the Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 2004;45(1):59–62.
- Landers JA, et al. Heritability of central corneal thickness in nuclear families. Invest Ophthalmol Vis Sci. 2009;50(9):4087–4090.
- Duggal P, Klein AP, Lee KE, Klein R, Klein BE, Bailey-Wilson JE. Identification of novel genetic loci for intraocular pressure: a genomewide scan of the Beaver Dam Eye Study. Arch Ophthalmol. 2007;125(1):74–79.
- Dimasi DP, et al. Novel quantitative trait loci for central corneal thickness identified by candidate gene analysis of osteogenesis imperfecta genes. Hum Genet. 2010;127(1):33–44.
- Macgregor S, et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum Mol Genet. 2010;19(13):2716–2724.View this article via: PubMed Google Scholar
- Mackey DA, Craig JE. Predictive DNA testing for glaucoma: reality in 2003. Ophthalmol Clin North Am. 2003;16(4):639–645.
- Fan BJ, Tam PO, Choy KW, Wang DY, Lam DS, Pang CP. Molecular diagnostics of genetic eye diseases. Clin Biochem. 2006;39(3):231–239.
- Golubnitschaja O, Flammer J. What are the biomarkers for glaucoma? Surv Ophthalmol. 2007;52(suppl 2):S155–S161.
- Dervan EW, et al. Protein macroarray profiling of serum autoantibodies in pseudoexfoliation glaucoma. Invest Ophthalmol Vis Sci. 2010;51(6):2968–2975.
- Honda N, et al. Effect of latanoprost on the expression of matrix metalloproteinases and tissue inhibitor of metalloproteinase 1 on the ocular surface. Arch Ophthalmol. 2010;128(4):466–471.
- Johnson EC, Guo Y, Cepurna WO, Morrison JC. Neurotrophin roles in retinal ganglion ell survival: lessons from rat glaucoma models. Exp Eye Res. 2009;88(4):808–815.
- Weber AJ, Viswanáthan S, Ramanathan C, Harman CD. Combined application of BDNF to the eye and brain enhances ganglion cell survival and function in the cat after optic nerve injury. Invest Ophthalmol Vis Sci. 2010;51(1):327–334.
- Martin KR, et al. Gene therapy with brain-derived neurotrophic factor as a protection: retinal ganglion cells in a rat glaucoma model. Invest Ophthalmol Vis Sci. 2003;44(10):4357–4365.
- Fu QL, et al. Combined effect of brain-derived neurotrophic factor and LINGO-1 fusion protein on long-term survival of retinal ganglion cells in chronic glaucoma. Neuroscience. 2009;162(2):375–382.
- Pease ME, et al. Effect of CNTF on retinal ganglion cell survival in experimental glaucoma. Invest Ophthalmol Vis Sci. 2009;50(5):2194–2200.
- Lambiase A, et al. Experimental and clinical evidence of neuroprotection by nerve growth factor eye drops: Implications for glaucoma. Proc Natl Acad Sci U S A. 2009;106(32):13469–13474.View this article via: PubMed Google Scholar
- Maguire AM, et al. Age-dependent effects of RPE65 gene therapy for Leber’s congenital amaurosis: a phase 1 dose-escalation trial. Lancet. 2009;374(9701):1597–1605.
- Liu X, Rasmussen CA, Gabelt BT, Brandt CR, Kaufman PL. Gene therapy targeting glaucoma: where are we? Surv Ophthalmol. 2009;54(4):472–486.
- Spiga MG, Borras T. Development of a gene therapy virus with a glucocorticoid-inducible MMP1 for the treatment of steroid glaucoma. Invest Ophthalmol Vis Sci. 2010;51(6):3029–3041.
- Barraza RA, McLaren JW, Poeschla EM. Prostaglandin pathway gene therapy for sustained reduction of intraocular pressure. Mol Ther. 2010;18(3):491–501.
- Buie LK, et al. Self-complementary AAV virus (scAAV) safe and long-term gene transfer in the trabecular meshwork of living rats and monkeys. Invest Ophthalmol Vis Sci. 2010;51(1):236–248.
- Bull ND, Johnson TV, Martin KR. Stem cells for neuroprotection in glaucoma. Prog Brain Res. 2008;173:511–519.View this article via: PubMed Google Scholar
- Parameswaran S, et al. Induced pluripotent stem cells (iPSCs) generate both retinal ganglion cells and photoreceptors: Therapeutic implications in degenerative changes in glaucoma and age-related macular degeneration. Stem Cells. 2010;28(4):695–703.
- Dahlmann-Noor AH, Vijay S, Limb GA, Khaw PT. Strategies for optic nerve rescue and regeneration in glaucoma and other optic neuropathies. Drug Discov Today. 2010;15(7–8):287–299.
- Johnson TV, Bull ND, Hunt DP, Marina N, Tomarev SI, Martin KR. Local mesenchymal stem cell transplantation confers neuroprotection in experimental glaucoma. Invest Ophthalmol Vis Sci. 2010;51:2051–2059.
- Bull ND, Irvine KA, Franklin RJ, Martin KR. Transplanted oligodendrocyte precursor cells reduce neurodegeneration in a model of glaucoma. Invest Ophthalmol Vis Sci. 2009;50(9):4244–4253.
- Kelley MJ, Rose AY, Keller KE, Hessle H, Samples JR, Acott TS. Stem cells in the trabecular meshwork: present and future promises. Exp Eye Res. 2009;88(4):747–751.
- Kass MA, et al. Delaying treatment of ocular hypertension: the ocular hypertension treatment study. Arch Ophthalmol. 2010;128(3):276–287.
- Ocular Hypertension Treatment Study Group, et al. Validated prediction model for the development of primary open-angle glaucoma in individuals with ocular hypertension. Ophthalmology. 2007;114(1):10–19.
- Leung CK, et al. Retinal nerve fiber layer imaging with spectral-domain optical coherence tomography a study on diagnostic agreement with Heidelberg Retinal Tomograph. Ophthalmology. 2010;117(2):267–274.
- Schmitz-Valckenberg S, et al. Real-time in vivo imaging of retinal cell apoptosis after laser exposure. Invest Ophthalmol Vis Sci. 2008;49(6):2773–2780.
- Suriyapperuma SP, et al. A new locus (GLC1H) for adult-onset primary open-angle glaucoma maps to the 2p15-p16 region. Arch Ophthalmol. 2007;125(1):86–92.
- Stoilova D, Child A, Trifan OC, Crick RP, Coakes RL, Sarfarazi M. Localization of a locus (GLC1B) for adult-onset primary open angle glaucoma to the 2cen-q13 region. Genomics. 1996;36(1):142–150.
- Sherwin JC, Hewitt AW, Bennett SL, Baird PN, Craig JE, Mackey DA. Primary open angle glaucoma in subjects harbouring the predicted GLC1L haplotype reveals a normotensive phenotype. Clin Experiment Ophthalmol. 2009;37(2):201–207.View this article via: PubMed Google Scholar
- Wirtz MK, et al. Mapping a gene for adult-onset primary open-angle glaucoma to chromosome 3q. Am J Hum Genet. 1997;60(2):296–304.View this article via: PubMed Google Scholar
- Wirtz MK, et al. GLC1F, a new primary open-angle glaucoma locus, maps to 7q35-q36. Arch Ophthalmol. 1999;117(2):237–241.View this article via: PubMed Google Scholar
- Trifan OC, et al. A third locus (GLC1D) for adult-onset primary open-angle glaucoma maps to the 8q23 region. Am J Ophthalmol. 1998;126(1):17–28.
- Phillips JC, et al. A second locus for Rieger syndrome maps to chromosome 13q14. Am J Hum Genet. 1996;59(3):613–619.View this article via: PubMed Google Scholar
- Inagaki Y, et al. Polymorphism of beta-adrenergic receptors and susceptibility to open-angle glaucoma. Mol Vis. 2006;12:673–680.View this article via: PubMed Google Scholar
- Lam CY, et al. Association of apolipoprotein E polymorphisms with normal tension glaucoma in a Chinese population. J Glaucoma. 2006;15(3):218–222.
- Fourgeux C, et al. Primary open-angle glaucoma: association with cholesterol 24S-hydroxylase (CYP46A1) gene polymorphism and plasma 24-hydroxycholesterol levels. Invest Ophthalmol Vis Sci. 2009;50(12):5712–5717.
- Kim SH, et al. Investigations on the association between normal tension glaucoma and single nucleotide polymorphisms of the endothelin-1 and endothelin receptor genes. Mol Vis. 2006;12:1016–1021.View this article via: PubMed Google Scholar
- Ayub H, et al. Association of eNOS and HSP70 gene polymorphisms with glaucoma in Pakistani cohorts. Mol Vis. 2010;16:18–25.View this article via: PubMed Google Scholar
- Tsai FJ, Lin HJ, Chen WC, Chen HY, Fan SS. Insulin-like growth factor-II gene polymorphism is associated with primary open angle glaucoma. J Clin Lab Anal. 2003;17(6):259–263.
- Clement CI, Goldberg I, Healey PR, Graham SL. Plasma homocysteine, MTHFR gene mutation, and open-angle glaucoma. J Glaucoma. 2009;18(1):73–78.
- Wolf C, et al. Evaluation of nine candidate genes in patients with normal tension glaucoma: a case control study. BMC Med Genet. 2009;10:91.
- Funayama T, et al. SNPs and interaction analyses of noelin 2, myocilin, and optineurin genes in Japanese patients with open-angle glaucoma. Invest Ophthalmol Vis Sci. 2006;47(12):5368–5375.
- Yu-Wai-Man P, et al. OPA1 increases the risk of normal but not high tension glaucoma. J Med Genet. 2010;47(2):120–125.
- Inagaki Y, et al. Paraoxonase 1 gene polymorphisms influence clinical features of open-angle glaucoma. Graefes Arch Clin Exp Ophthalmol. 2006;244(8):984–990.View this article via: PubMed Google Scholar
- Lin HJ, Tsai CH, Tsai FJ, Chen WC, Chen HY, Fan SS. Transporter associated with antigen processing gene 1 codon 333 and codon 637 polymorphisms are associated with primary open-angle glaucoma. Mol Diagn. 2004;8(4):245–252.View this article via: PubMed Google Scholar
- Shibuya E, et al. Association of Toll-like receptor 4 gene polymorphisms with normal tension glaucoma. Invest Ophthalmol Vis Sci. 2008;49(10):4453–4457.
-
Fan BJ, et al. Association of polymorphisms of tumor necrosis factor and tumor protein p53 with primary open angle glaucoma: A replication study of ten genes in a Chinese population [published online ahead of print March 31, 2010]. Invest Ophthalmol Vis Sci
. doi:10.1167/iovs.09-4974
. View this article via: CrossRef Google Scholar
-
Version history
- Version 1 (September 1, 2010): No description
Article tools
-
View PDF

- Download citation information
- Send a comment
- Terms of use
- Standard abbreviations
- Need help? Email the journal
Review Series
The Eye
-
Retinopathy of prematurity: understanding ischemic retinal vasculopathies at an extreme of life
Przemyslaw Sapieha et al. -
How cortical neurons help us see: visual recognition in the human brain
Julie Blumberg et al. -
Lighting a candle in the dark: advances in genetics and gene therapy of recessive retinal dystrophies
Anneke I. den Hollander et al. -
Therapeutic targets in age-related macular disease
Alan C. Bird -
A look at autoimmunity and inflammation in the eye
Rachel R. Caspi -
Glaucoma: genes, phenotypes, and new directions for therapy
Bao Jian Fan et al. -
Stemming vision loss with stem cells
Valentina Marchetti et al. -
An eye for discovery
Andreas Stahl et al.
Metrics
Go to
- Top
- Abstract
- Introduction
- Genes identified using linkage approaches and multiplex families
- Case-control association studies: POAG and NTG
- Genome-wide association studies
- Pathophysiology associated with current glaucoma genes
- Quantitative traits
- Biomarkers for glaucoma
- Experimental therapeutic approaches
- Future directions
- Acknowledgments
- Footnotes
- References
- Version history



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)




