Advertisement
Research ArticleGastroenterologyImmunology
Open Access |  10.1172/JCI187499
10.1172/JCI187499
A20’s linear ubiquitin–binding motif restrains pathogenic activation of Th17 cells and IL-22–driven enteritis
Christopher J. Bowman,1,2 Dorothea M. Stibor,1 Xiaofei Sun,1 Nika Lenci,1 Hiromichi Shimizu,1 Emily F. Yamashita,1 Rommel Advincula,1 Min Cheol Kim,3 Jessie A. Turnbaugh,4 Yang Sun,3 Bahram Razani,5 Peter J. Turnbaugh,4 Chun Jimmie Ye,1,3,6 Barbara A. Malynn,1 and Averil Ma1
1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by Bowman, C. in: PubMed | Google Scholar
1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by Stibor, D. in: PubMed | Google Scholar
1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by Sun, X. in: PubMed | Google Scholar
1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by Lenci, N. in: PubMed | Google Scholar
1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by Shimizu, H. in: PubMed | Google Scholar
1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by Yamashita, E. in: PubMed | Google Scholar
1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by Advincula, R. in: PubMed | Google Scholar
1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by Kim, M. in: PubMed | Google Scholar
1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by Turnbaugh, J. in: PubMed | Google Scholar
1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by Sun, Y. in: PubMed | Google Scholar
1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by
Razani, B.
in:
PubMed
|
Google Scholar
|

1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by Turnbaugh, P. in: PubMed | Google Scholar
1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by Ye, C. in: PubMed | Google Scholar
1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by
Malynn, B.
in:
PubMed
|
Google Scholar
|

1Department of Medicine,
2Department of Pathology,
3Department of Epidemiology and Biostatistics,
4Department of Microbiology and Immunology, and
5Department of Dermatology, University of California, San Francisco (UCSF), San Francisco, California, USA.
6Gladstone-UCSF Institute of Genomic Immunology, San Francisco, California, USA.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
Authorship note: CJB, DS, and XS have been designated as co–first authors.
Find articles by
Ma, A.
in:
PubMed
|
Google Scholar
|

Authorship note: CJB, DS, and XS have been designated as co–first authors.
Published September 2, 2025 - More info
J Clin Invest. 2025;135(17):e187499. https://doi.org/10.1172/JCI187499.
© 2025 Bowman et al. This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Received: October 1, 2024; Accepted: June 17, 2025
-
Abstract
A20, encoded by the TNFAIP3 gene, is a protein linked to Crohn’s disease and celiac disease in humans. We now find that mice expressing point mutations in A20’s M1-ubiquitin–binding zinc finger 7 (ZF7) motif spontaneously develop proximal enteritis that requires both luminal microbes and T cells. Cellular and transcriptomic profiling reveals expansion of Th17 cells and exuberant expression of IL-17A and IL-22 in intestinal lamina propria of A20ZF7 mice. While deletion of IL-17A from A20ZF7/ZF7 mice exacerbates enteritis, deletion of IL-22 abrogates intestinal epithelial cell hyperproliferation, barrier dysfunction, and alarmin expression. Colonization of adult germ-free mice with microbiota from adult WT specific pathogen–free mice drives duodenal IL-22 expression and duodenitis. A20ZF7/ZF7 Th17 cells autonomously express more RORγt and IL-22 after differentiation in vitro. ATAC sequencing identified an enhancer region upstream of the Il22 gene, and this enhancer demonstrated increased activating histone acetylation coupled with exaggerated Il22 transcription in A20ZF7/ZF7 T cells. Acute inhibition of RORγt normalized histone acetylation at this enhancer. Finally, CRISPR/Cas9–mediated ablation of A20ZF7 in human T cells increases RORγt expression and IL22 transcription. These studies link A20’s M1-ubiquitin binding function with RORγt expression, expansion of Th17 cells, and epigenetic activation of IL-22–driven enteritis.
-
Introduction
Intestinal immune homeostasis involves complex interactions between immune and non-immune cells. Many of these interactions are mediated by cytokines, and disruption of this cytokine network can lead to intestinal disease. Type 3 cytokines such as IL-17A, IL-17F, and IL-22 support antifungal and antibacterial responses (1). These mediators can also support epithelial homeostasis and regeneration (2). Dysregulated expression of type 3 cytokines is a feature of intestinal inflammation, and expansion of Th17 cells has been consistently observed in experimental models and human inflammatory bowel disease (IBD) patients (3, 4). However, IL-17A appears to play more complex roles in the intestinal milieu than in other tissues (5, 6). Some of this complexity may be related to the ability of Th17 cells to produce other cytokines under distinct physiological conditions. Hence, understanding the physiological regulation of intestinal cytokines and cytokine responses is crucial for dissecting the adaptive versus pathological functions of these cytokine mediators.
A20/TNFAIP3 is genetically linked to IBD and celiac disease via GWAS (7–9). In addition, rare patients harboring haploinsufficient mutations of the A20 gene develop early-onset IBD as well as Behçet’s disease with intestinal ulcerations (10–12). Hence, deficiencies of A20 expression and/or function likely compromise human intestinal homeostasis. Mechanistic studies have revealed that the A20 protein regulates several signaling cascades, including TNFR-, TLR-, TCR-, NOD2-, and CD40R-triggered signals (13–19). A20 performs these functions by regulating the ubiquitination of critical signaling molecules such as RIP1, RIP2, pro–IL-1β, and RIP3 as well as ubiquitinated signaling complexes such as the IKKγ complex. Yet the mechanisms by which A20 preserves intestinal immunity are incompletely understood. This is partly because the A20 protein harbors distinct biochemical domains that mediate deubiquitinating (DUB), E3 ubiquitin (Ub) ligase, and non-catalytic linear (M1)-Ub chain binding activities (13, 14, 20–25). In this study, we unveil a unique role for A20’s M1-Ub–binding motif in restraining epigenetic regulation of IL-22 expression in Th17 cells, intestinal epithelial cell homeostasis, and microbe-dependent enteritis.
-
Results
Linear (M1)-ubiquitin–binding motif of A20 prevents T cell–dependent enteritis. To understand the biochemical functions of A20 that regulate intestinal homeostasis, we analyzed intestines from a series of A20 knock-in mice that abrogate A20’s DUB (A20OTU), E3 Ub ligase (A20ZF4), or M1-Ub binding (A20ZF7) activities (20, 21). Macroscopically, small intestines from 12-week-old A20ZF7/ZF7 mice, but not other genotypes, demonstrated visible thickening of the intestinal wall. Histology suggested that the proximal small intestinal mucosa of A20ZF7/ZF7 mice harbored increased numbers of immune cells in comparison with congenic A20ZF4/ZF4, A20OTU/OTU, or wild-type (WT) mice (Figure 1, A and B). The expansion of the small intestinal lamina propria (SILP) by lymphocytes is reminiscent of chronic enteritis; however, other features of chronicity (e.g., epithelial metaplasia, villus blunting) were absent. This phenotype was evident in both male and female A20ZF7/ZF7 mice and was 100% penetrant by 12 weeks of age. This phenotype was not evident in more distal portions of the small intestine and large intestines from 12-week-old A20ZF7/ZF7 mice. In particular, ileal and colonic tissues from A20ZF7/ZF7 mice expressed normal histology without molecular markers of inflammation (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI187499DS1). The selective enhanced inflammation of proximal small intestines of A20ZF7/ZF7 mice differs from other spontaneous models of intestinal inflammation such as Il2–/–, Il10–/–, and TnfΔARE mice. Hence, A20’s M1-Ub binding activity via its zinc finger 7 (ZF7) domain preserves small intestinal immune homeostasis independently of A20’s DUB and E3 Ub ligase activities.
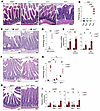 Figure 1
Figure 1A20ZF7 restrains small intestinal enteritis that is T cell dependent. (A and B) Representative H&E histology (A) and histopathological scores (B) of inflammation of proximal small intestines from indicated genotypes of mice. Scale bars: 200 μm. (C and D) Representative H&E histology (C) and histopathological scores (D) of inflammation of small intestine from radiation bone marrow chimeric mice reconstituted with indicated bone marrow genotypes. Scale bars: 100 μm. (E) Quantitative PCR (qPCR) analyses of indicated proinflammatory genes in proximal small intestines from indicated bone marrow chimeras. (F) Representative H&E histology of small intestine from indicated genotypes of Rag1–/– mice. Scale bars: 100 μm. Data are representative of at least 4 independent pairs of mice. (G) Representative H&E histology of small intestine from indicated genotypes of Ighm–/– mice. Scale bars: 100 μm. Data are representative of at least 3 independent pairs of mice. (H) Pathology scores of small intestinal inflammation. (I) qPCR analyses of indicated proinflammatory genes in proximal small intestines from indicated genotypes of mice. In panels above, each circle represents 1 mouse. Data are shown as mean ± SEM. Statistics were calculated using non-parametric Kruskal-Wallis test with Dunn’s multiple-comparison correction (B), unpaired 2-tailed Mann-Whitney U test (D and H), or unpaired 2-tailed Student’s t test with Welch’s correction (E and I). *P < 0.05, **P < 0.01, ***P < 0.001.
A20 is expressed and induced in many cell types, including both hematopoietic and non-hematopoietic cells (26). To determine the degree to which hematopoietic cells of A20ZF7/ZF7 mice (hereafter designated A20ZF7 mice) are sufficient to drive this pathology, we generated radiation chimeras using either A20ZF7 or WT bone marrow cells to reconstitute irradiated WT mice Chimeras bearing A20ZF7 hematopoietic cells spontaneously developed enteritis by 12 weeks after reconstitution, a phenotype that was not seen in chimeras containing WT hematopoietic cells (Figure 1, C and D). Furthermore, the A20ZF7 hematopoietic cells drove exaggerated expression of proinflammatory myeloid cytokines such as Il1b and chemokines such as Ccl20 (Figure 1E). To assess the relative contributions of T and B cells to enteritis in A20ZF7 mice, we interbred these mice with Rag1–/– and Ighm–/– mice. Twelve-week-old A20ZF7 Rag1–/– mice exhibited negligible intestinal inflammation and expressed levels of Il1b and Ccl20 similar to those in Rag1–/– mice (Figure 1, F, H, and I), suggesting that adaptive lymphocytes are required for enteritis in A20ZF7 mice. By contrast, intestines from A20ZF7 Ighm–/– mice accumulated similar numbers of lamina propria immune cells (Figure 1, G and H) and expressed similarly elevated levels of inflammatory markers in comparison with A20ZF7 mice (Figure 1I). Hence, Rag1-dependent T lymphocytes but not B lymphocytes are required for small intestinal inflammation in A20ZF7 mice.
Type 3 cytokines and Th17 cells are increased in intestinal lamina propria of A20ZF7 mice. To better understand why small intestinal inflammation develops in A20ZF7 mice, we analyzed the transcriptomes of intact small intestines from 12-week-old WT and A20ZF7 mice by bulk RNA sequencing (RNA-Seq). These studies revealed that A20ZF7 mice upregulated genes involved in NF-κB and inflammasome signaling pathways (Figure 2A). In addition, A20ZF7 intestines expressed elevated levels of genes associated with CD4+ T cell activation, T cell proliferation, and IL-1β production (Figure 2A). IL-17 response genes were among the most enriched groups of genes, which — together with STAT and IL-6 regulation — suggests a prominent type 3 cytokine tone in A20ZF7 intestines. Indeed, quantitative mRNA analyses of intact intestines confirmed the exaggerated expression of type 3 cytokines, Il17a and Il22 (Figure 2B). Meanwhile, Il17ra levels were decreased in A20ZF7 intestines, an expected downregulation in response to elevated IL-17 (27). Surprisingly, expression of proinflammatory Il23a and Il18 was diminished and Il23r was not significantly elevated in A20ZF7 intestines (Figure 2B). IL-22–dependent transcripts, such as Reg3b, Reg3g, Saa1, and Saa3, were significantly upregulated in A20ZF7 intestines (Figure 2C and Figure 5E), while expression of Il22ra2 was not significantly altered (Figure 2B). As these data suggest that Th17 cells and/or group 3 innate lymphoid cells (ILC3s) might be expanded or hyperfunctional in A20ZF7 intestines, we profiled cellular infiltrates from small intestinal tissues. Immunohistochemistry of intact small intestines indicated that CD4+ T cells were expanded in the lamina propria of A20ZF7 mice (Figure 2D), and flow cytometry of dissociated SILP cells confirmed a relative expansion of CD4+ T cells (Figure 2E). The increased number of CD4+ T cells in A20ZF7 SILP largely comprised an expansion of Th17 cells in comparison with WT littermates (Figure 2E). In addition, among expanded A20ZF7 SILP CD4+ T cells, IL-17A– and IL-17F–expressing cells were disproportionally increased while IFN-γ–expressing cells were present in similar proportions to those of WT mice.
 Figure 2
Figure 2Small intestines of A20ZF7 mice harbor expanded Th17 cells. (A) Gene ontology enrichment of genes that are significantly upregulated in A20ZF7 versus WT intestines by bulk RNA-Seq. Red bars highlight categories related to Th17 differentiation. Blue bars highlight categories related to cellular proliferation. (B) qPCR analyses of mRNA expression from intact small intestine. (C) Volcano plot of all annotated UCSC RefSeq genes from bulk RNA-Seq analyses of A20ZF7 versus WT small intestines. Horizontal dotted line indicates adjusted P value (FDR) of 0.01. (D) Representative immunohistochemical analyses of CD4 expression in WT and A20ZF7 small intestines. Data are representative of 3 mice from each genotype. Scale bars: 100 μm. (E) Flow cytometry of small intestinal lamina propria (SILP) cells from WT and A20ZF7 mice. SILP yields from the proximal 10 cm of small intestine typically range from 1 million to 2 million cells from WT mice and 3 million to 7 million cells from A20ZF7 mice. Data are shown as mean ± SEM. Statistics were calculated using unpaired 2-tailed Student’s t test with Welch’s correction. *P < 0.05, **P < 0.01, ***P < 0.001.
To better define the SILP lymphocytes in A20ZF7 mice, we enriched SILP T cells from 12-week-old WT and A20ZF7 mice and profiled these cells by single-cell RNA-Seq (scRNA-Seq). To avoid clustering cells based on cell cycle phases, genes related to cell cycling were removed before clustering analyses. Uniform manifold approximation and projection (UMAP) analyses of these cell cycle–regressed cells identified CD8+, Th17, and regulatory T cells, as well as subsets of innate lymphoid cells (ILCs) (Figure 3A and Supplemental Figure 2A). Genotype-specific UMAP analyses showed a relative expansion of CD4+ and, to a lesser extent, CD8+ T cells in A20ZF7 mice (Figure 3, B and C).
 Figure 3
Figure 3A20ZF7 mice show proliferation and expansion of Th17 and regulatory T cells within the SILP. (A and B) UMAP clusters of scRNA-Seq analyses of SILP from WT and A20ZF7 mice. Data represent pooled mixtures of 2 WT and 2 A20ZF7 mice. (C) Relative proportions of lymphoid subsets in WT and A20ZF7 SILP. Th17 subset includes both proliferative (mustard yellow) and non-proliferative (sky blue) compartments. (D) Projection of Il17a and Il22 expression onto UMAP clusters shown in B. (E) Violin plots of Il17a and Il22 expression in Th17 cells from indicated genotypes of mice. Statistics were calculated using unpaired 2-tailed Wilcoxon’s rank sum test. **P < 0.01, ***P < 0.001.
Expanded clusters of CD4+ T cells in A20ZF7 intestines contained many cells expressing Il17a and Il22, delineating these cells as Th17 cells (Figure 3D and Supplemental Figure 2A). Another expanded cluster included regulatory T cells (Figure 3C). By contrast, ILC3s were relatively reduced in A20ZF7 mice (Figure 3C). In addition to being more abundant in A20ZF7 mice, A20ZF7 Th17 cells expressed higher levels of Il17a and Il22 than WT Th17 cells (Figure 3E). The marked expansion of Th17 cells, the increased expression of type 3 cytokines, and the absence of ILC3 expansion in A20ZF7 intestines relative to WT intestines suggest that Th17 cells account for the great majority of the increased type 3 cytokine production in A20ZF7 intestines.
The Th17 cells segregated into 2 discrete clusters in UMAP space, and both clusters were heavily represented in A20ZF7 intestines (Figure 3, A and B, and Supplemental Figure 2A). After the contributions of cell cycle genes had been removed as an effect on cell clustering, these two clusters continued to exhibit differential cell cycle states: cells with high G0/G1 scores (indicating a predominantly non-proliferative state) and cells enriched for high G2/M or S scores (suggesting T cell proliferation) (Supplemental Figure 2B). In addition, this “Th17 proliferative” cluster expressed high levels of Mki67, the gene that encodes the proliferative marker Ki67 (Supplemental Figure 2A). The accumulation of proliferative Th17 cells in A20ZF7 SILP aligns with our bulk RNA-Seq analysis, which highlighted T cell proliferation as an enriched category in A20ZF7 intestines (Figure 2A). Although the proliferative cluster was transcriptionally distinct from non-proliferating T cells in either A20ZF7 or WT mice, it consisted of cells that expressed detectable levels of Il17a and/or Il22, confirming they were Th17 cells (Figure 3D and Supplemental Figure 2A). Hence, increased numbers of proliferating Th17 cells help explain why Th17 cells are more numerous in A20ZF7 intestines.
IL-22, but not IL-17A, drives enteritis in A20ZF7 mice. As A20ZF7 mouse small intestines contain dramatic expansions of SILP Th17 cells and increased tissue-wide expression of IL-17A and IL-22, we next sought to functionally define the potential roles of these cytokines in regulating intestinal disease in A20ZF7 mice. We interbred A20ZF7 mice with Il17a–/– mice and Il22–/– mice. Small intestines from A20ZF7 Il17a–/– mice exhibited more (rather than less) severe enteritis than those from IL-17A–competent A20ZF7 mice (Figure 4, A and B). Hence, IL-17A plays a protective rather than proinflammatory role in enteritis in A20ZF7 mice. A20ZF7 Il17a–/– intestines expressed more Il22 than Il17a–/– intestines but not A20ZF7 intestines (Figure 4C). In contrast to A20ZF7 Il17a–/– mice, A20ZF7 Il22–/– mice exhibited less severe enteritis than A20ZF7 mice (Figure 4, A and B). Therefore, IL-22 promotes small intestinal inflammation in A20ZF7 mice. To better define the role of IL-22 in A20ZF7 intestines, we profiled the genome-wide transcriptomes of small intestines from A20ZF7 Il22–/– and control mice by bulk RNA-Seq. Principal component analysis of bulk RNA-Seq data revealed broad normalization toward wild-type transcriptomic states in A20ZF7 Il22–/– mice when compared with A20ZF7 mice (Figure 4D). Thus, IL-22 drives transcriptome-wide changes in the proximal small intestine that promote intestinal inflammation in A20ZF7 mice.
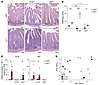 Figure 4
Figure 4IL-17A protects against and IL-22 promotes small intestinal enteritis in A20ZF7 mice. (A and B) Representative H&E histology (A) and pathology scores (B) of small intestines from mice of the indicated genotypes. Scale bars: 100 μm. (C) qPCR analyses of type 3 cytokines in small intestines from indicated genotypes of mice. (D) Principal component analyses of bulk RNA-Seq analyses of indicated genotypes of mice. Data are shown as mean ± SEM. Statistics were calculated using 2-way ANOVA with Holm-Šidák multiple-comparison correction (B) or unpaired 2-tailed Student’s t test with Welch’s correction (C). *P < 0.05, **P < 0.01, ***P < 0.001.
A20ZF7 mice exhibit IL-22– and microbe-dependent epithelial barrier dysfunction. Il22ra1, the IL-22–specific receptor chain, is selectively expressed on intestinal epithelial cells (IECs) but not immune cells (28). Hence, pathophysiological effects of IL-22 in A20ZF7 mice could be mediated by perturbation of IEC functions. By histology, epithelial crypts were disproportionally expanded in small intestines of A20ZF7 mice when compared with WT mice (Figure 4A and Figure 5A). Bulk RNA-Seq highlighted epithelial cell proliferation as an enriched gene set in A20ZF7 intestines (Figure 2A), and immunohistochemistry for Ki67 confirmed an expansion of proliferating crypt IECs in A20ZF7 intestines (Figure 5, A and B). Interestingly, chimeras bearing A20ZF7 hematopoietic cells also expressed elevated levels of IEC-derived defensins Reg3b and Reg3g (Figure 5C). This result supports that radiation-sensitive A20ZF7 Th17 cells express exaggerated amounts of IL-22 that perturb WT IECs. Notably, the IEC proliferation and crypt elongation seen in A20ZF7 mice were significantly lessened in A20ZF7 Il22–/– mice (Figure 5, A and B). To understand the IL-22–dependent perturbations of IECs in A20ZF7 mice, we isolated small intestinal IECs from A20ZF7 Il22–/– and control mice. Transcriptomic analyses of these cells revealed that A20ZF7 epithelia expressed more C-type lectins (Reg3b, Reg3g), alarmins (Saa1), and chemokines (Cxcl1) than WT IECs (Figure 5, D and E). These defects were markedly reduced in A20ZF7 Il22–/– IECs, indicating that IL-22 drives exaggerated expression of these proinflammatory mediators in A20ZF7 IECs.
 Figure 5
Figure 5IL-22 disrupts epithelial barrier integrity and drives microbe-dependent enteritis in A20ZF7 mice. (A) Average crypt length measured in histology sections of proximal small intestines from indicated genotypes of mice. (B) Representative immunohistochemistry of Ki67 expression in small intestines of indicated genotypes of mice. Data are representative of 2–3 mice from each genotype. Scale bars: 100 μm. (C) qPCR analyses of IL-22–dependent defensins from intact small intestines of indicated radiation chimeras. (D) qPCR analyses of IL-22–dependent genes from isolated small intestinal IECs. (E) Normalized mRNA counts of IL-22–dependent genes from bulk RNA-Seq of intact small intestines. (F) Fluorescence measurements of FITC-dextran in sera from indicated genotypes of mice 4 hours after FITC-dextran gavage. (G) qPCR analyses of indicated tight junction genes from isolated small intestinal IECs from indicated genotypes of mice. (H) Representative H&E histology of proximal small intestines from germ-free (GF) or conventionalized (Conv.) mice of indicated genotypes. Scale bars: 200 μm. (I and J) qPCR analyses of indicated genes from small intestines from indicated genotypes of specific pathogen–free (SPF) and GF mice. (K) qPCR analyses of indicated genes from small intestines from indicated genotypes of GF or Conv. mice. Data are shown as mean ± SEM. Each circle represents an independent mouse. Statistics were calculated using unpaired 2-tailed Student’s t test with Welch’s correction (C–F, Il22 in K) or 2-way ANOVA with Bonferroni’s multiple-comparison correction (A, Ccl20 in E, G, I, J, Reg3b and Reg3g in K). *P < 0.05, **P < 0.01, ***P < 0.001.
As proinflammatory cytokines can perturb epithelial barrier functions (29–31), we tested intestinal epithelial permeability in A20ZF7 Il22–/– mice and control mice by gavaging these mice with FITC-dextran. Increased absorption of FITC-dextran in sera of A20ZF7 mice supports aberrant barrier integrity in these mice. This compromised barrier integrity was similarly exacerbated in A20ZF7 Il17a–/– mice but normalized in A20ZF7 Il22–/– mice (Figure 5F). To understand why A20ZF7 IECs fail to maintain barrier integrity, we assayed expression of occludin (Ocln) and claudin-4 (Cldn4), 2 tight junction proteins that support epithelial barrier integrity (32, 33). Ocln and Cldn4 expression were depressed in A20ZF7 compared with WT IECs, and Cldn4 was normalized in A20ZF7 Il22–/– IECs (Figure 5G). Although Ocln in A20ZF7 Il22–/– IECs did not reach levels similar to those in Il22–/– IECs, these were normalized to WT levels. Diminished barrier integrity may allow translocation of microbes and pathogenic products, inciting an inflammatory response (34–36). To determine whether intestinal microbiota are required for enteritis in A20ZF7 mice, these mice were derived into germ-free environments. Germ-free A20ZF7 mice exhibited neither intestinal inflammation (Figure 5H) nor elevated expression of proinflammatory cytokines (i.e., Il1b, Ccl20, Tnf, Ifng) relative to controls (Figure 5I), a stark contrast to the A20ZF7 mice raised in specific pathogen–free (SPF) environments. Despite the mutation of A20ZF7 in the small intestines of germ-free mice, the absence of commensal microbiota did not lead to alarmin (Saa1, Saa3) production (Figure 5J), suggesting that microbiota are necessary for IL-22–dependent programs within the small intestine. Segmented filamentous bacteria (SFB) preferentially colonize and induce Th17 responses in the terminal ileum but not duodenum (37–40). Specific PCR detection of SFB 16S rRNA in our mice infrequently showed SFB in ilea of WT and A20ZF7 intestines (Supplemental Figure 3A) and never detected SFB in duodena from either genotype. Therefore, the duodenitis of A20ZF7 mice is microbe dependent but unlikely to be driven by SFB.
As our SPF A20ZF7 and WT mice were cohoused, thereby sharing luminal microbes via coprophagic behavior, A20ZF7 intestines may have responded aberrantly to the same commensal microbes that were present in WT cagemates. To further test this notion, we harvested luminal commensal microbes from an unrelated colony of WT C57BL/6 mice, pooled these organisms, and introduced these microbes (“WT” microbes) into cohoused 6-week-old A20ZF7 and WT germ-free mice. These neocolonized, or “conventionalized,” mice were maintained as cagemates for 6 weeks. Analyses of intestines from these mice revealed that duodenal tissues from conventionalized WT mice minimally expressed Il22 and IL-22–dependent genes compared with those from germ-free WT mice (Figure 5K). By contrast, conventionalized A20ZF7 mice expressed markedly elevated levels of these genes compared with germ-free A20ZF7 mice or conventionalized WT mice (Figure 5K). Correspondingly, conventionalized A20ZF7 mice exhibited marked duodenal, but not ileal or colonic, inflammation when compared with conventionalized WT mice (Figure 5H and Supplemental Figure 1D). Moreover, germ-free and conventionalized mice harbored no detectable SFB (Supplemental Figure 3B), further supporting that the duodenal inflammation in A20ZF7 mice is microbe dependent but, also, independent of SFB. Taken together, these data indicate that A20ZF7 intestines respond aberrantly to “WT” microbiota by expressing markedly elevated expression of IL-22 that, in turn, perturbs IEC functions, disrupts intestinal epithelial barrier integrity, and causes duodenitis.
A20ZF7 restrains IL-22 expression in murine and human CD4+ T cells. Our data above show that T cell–autonomous A20ZF7 functions and IL-22 are integral to enteritis in A20ZF7 mice. Accordingly, we investigated how A20ZF7 regulates T cell production of IL-22. We enriched naive CD4+ T cells from 8-week-old A20ZF7 and WT mice and differentiated these cells into Th17 cells with recombinant IL-6 and TGF-β. The efficiency of in vitro Th17 differentiation (as defined by the expression of RORγt, the key transcription factor that coordinates Th17 differentiation) (41) was consistently greater than 99% (Figure 6A), regardless of A20 mutation status. Notably, A20ZF7 Th17 cells exhibited greater amounts of RORγt localized to the nucleus than WT Th17 cells, suggesting that A20ZF7 restrains RORγt expression and/or nuclear localization in a cell-autonomous fashion (Figure 6A). A20ZF7 Th17 cells also expressed more phosphorylated Stat3, reflecting substantial activation of this pathway (Figure 6B). In the absence of PMA/ionomycin stimulation, A20ZF7 Th17 cells also expressed more Il22 mRNA than WT cells (Figure 6C). Since Il22 expression is dependent on the aryl hydrocarbon receptor (Ahr) (42), these cells were treated with the Ahr agonist FICZ (42, 43). FICZ further exaggerated enhanced Il22 expression in A20ZF7 Th17 cells relative to WT cells (Figure 6C). Similarly, ELISA of supernatants from these cells revealed that A20ZF7 T cells secreted more IL-22 protein than WT cells (Figure 6D). Notably, these results were obtained from cells that were not stimulated with PMA or ionomycin, avoiding potential caveats associated with possible A20-dependent regulation of these stimuli. Aligned with these findings, flow cytometry studies of PMA/ionomycin-stimulated cells showed increased numbers of IL-22+ T cells in A20ZF7 cultures when compared with WT cultures (Supplemental Figure 4B). Hence, mutation of A20ZF7 within T cells leads to greater RORγt expression and increased expression of IL-22.
 Figure 6
Figure 6A20ZF7 Th17 cells show enhanced IL-22 production due to increased histone acetylation at an Il22 enhancer. (A and B) Flow nucleometry of RORγt (A) or phosphorylated Stat3 (Tyr705) (B) expression in nuclei isolated from in vitro–differentiated Th17 cells from WT and A20ZF7 mice. Histograms are representative of n = 2–3 biologically independent experiments. (C) qPCR analyses of Il17a and Il22 expression in cells generated as in A. “Th17” indicates Th17 differentiation conditions. “FICZ” indicates treatment with Ahr agonist FICZ. (D) ELISA of IL-22 secretion from cells generated as in A. (E) ATAC-seq of genomic loci at or near the Il22 locus in cells generated as in A. (F) ChIP of acetylated H3K27 at indicated Il22 loci (loci a–c in E) in cells generated as in A. (G) ChIP of histone H3 at the Il22 promoter and enhancer loci (loci a–c in E). Cells were generated as in A and treated with the RORγt inhibitor GSK805. H3 ChIP directly assesses nucleosome occupancy and, thus, chromatin accessibility. (H) ChIP of acetylated H3K27 at Il22 enhancer loci (loci a–c in E). Cells were generated as in A and treated with the RORγt inhibitor GSK805. (I) Flow cytometric analyses of RORγt expression in CRISPR/Cas9–edited primary human T cells differentiated in vitro using Th17 conditions. (J) qPCR analyses of expression of indicated genes in paired isogenic human Th17 cells engineered with CRISPR/Cas9 and either A20ZF7-targeted or nontargeting guide RNAs. Three pairs of isogenic samples from 2 healthy donors are shown. Data are shown as mean ± SEM. Statistics were calculated using unpaired 2-tailed Student’s t test with Welch’s correction (C, D, and F), 2-way ANOVA with post hoc Tukey’s multiple-comparison correction with simple effects (G and H), or paired-ratio t test (J). *P < 0.05, **P < 0.01, ***P < 0.001.
As A20ZF7 Th17 cells express more Il22 mRNA and protein than WT cells, we hypothesized that A20ZF7 restrains epigenetic regulation of Il22 transcription. To identify potential genomic loci that may regulate Il22 in cis, we surveyed chromatin accessibility via assay for transposase-accessible chromatin sequencing (ATAC-seq) followed by massively-parallel sequencing of Th17 cells differentiated from naive WT and A20ZF7 CD4+ T cells. ATAC-seq revealed increased accessibility across the Il22 gene in A20ZF7 Th17 cells (Figure 6E, region g), supporting open chromatin at Il22’s promoter and enhanced Il22 transcription. In addition, multiple other sites upstream of Il22 demonstrated DNA accessibility, predicting potential enhancer regions (Figure 6E, regions a–f). Notably, ATAC-seq highlighted a conserved region 32 kb upstream of Il22’s transcription start site (Figure 6E, region b), a site recently characterized as an Il22 enhancer (44). To define this conserved region as a bona fide functional enhancer and to quantify the activation state of this enhancer, naive WT and A20ZF7 CD4+ T cells were differentiated into Th17 cells, and chromatin immunoprecipitation (ChIP) of acetylated lysine 27 of histone H3 (H3K27ac) was performed at these genomic loci. H3K27ac was increased in A20ZF7 Th17 cells at the enhancer for Il22 (Figure 6F, region b), supporting increased enhancer activation and enhanced Il22 transcription in A20ZF7 Th17 cells. By contrast, sequences located on the immediate shoulders of the enhancer (Figure 6F, regions a and c) as well as other DNA accessible sites (Supplemental Figure 4C, regions d–f) were less decorated with H3K27ac and similarly marked in WT and A20ZF7 Th17 cells. The enhancement of H3K27ac marks at the conserved enhancer with FICZ treatment suggests that Ahr activation facilitates the acetylation of this locus. These results provide a molecular underpinning for Ahr in promoting Il22 expression (42, 43). As our flow studies of isolated nuclei showed that A20ZF7 Th17 cells harbor increased accumulation of nuclear RORγt compared with WT Th17 cells (Figure 6A), we next tested whether this increased RORγt mediates increased transcriptional activity at the Il22 enhancer. We differentiated naive CD4+ WT and A20ZF7 cells into Th17 cells in vitro and acutely treated cells with GSK805 (a RORγt-specific inhibitor) (45, 46) overnight beginning on day 6 of differentiation. Subsequent chromatin analyses at the enhancer showed that RORγt inhibition in A20ZF7 Th17 cells normalized nucleosome occupancy/density (Figure 6G) and H3K27ac (Figure 6H) to WT levels. These findings support RORγt’s role in reducing nucleosomal occupancy, facilitating chromatin and DNA accessibility, and enhancing acetylation of H3K27 at the Il22 enhancer in A20ZF7 Th17 cells. Therefore, elevated RORγt in differentiated A20ZF7 Th17 cells actively remodels chromatin at the Il22 enhancer to drive increased Il22 transcription. Taken together, these studies suggest that A20ZF7 restrains RORγt activity to limit chromatin accessibility and hyperacetylation of a specific Il22 enhancer, thus restricting Il22 transcription in CD4+ Th17 cells.
The TNFAIP3 gene is well conserved between murine and human genomes, and A20 polymorphisms and mutations are associated with human IBD and celiac disease (7, 10, 47). To determine whether A20’s ZF7 domain regulates Th17 cell functions and IL-22 expression in human T cells, we generated A20ZF7 mutant human CD4+ T cells using CRISPR/Cas9. Since A20’s ZF7 domain comprises the most C-terminal residues of the A20 protein, targeting A20ZF7 is unlikely to affect the stability or structure of the A20 protein. Our recent studies of murine A20ZF7 mutant proteins suggest that these proteins are indeed expressed at supranormal levels in TNF-stimulated fibroblasts (21). We thus designed CRISPR guide RNAs (gRNAs) to delete the N-terminal half of A20’s ZF7 domain. We isolated naive CD4+ T cells from healthy donor peripheral blood, activated the cells via TCR stimulation, and electroporated CRISPR/Cas9 along with either these ZF7-targeting or control gRNAs to generate A20ZF7 mutant and isogenic control human T cells. Sanger sequencing demonstrated that more than 85% of alleles were targeted by A20ZF7-specific gRNAs (Supplemental Figure 5, A and B). These cells were then differentiated toward Th17 cells for 9 days, at which time more than 99% expressed RORγt in both A20ZF7-mutated and control cells (Figure 6I), confirming efficient Th17 differentiation. Expression of Tnfaip3 mRNA is markedly induced by NF-κB activity, and A20 mediates negative feedback on NF-κB signaling (26). In keeping with the compromised ability of A20ZF7 cells to restrain NF-κB signaling, expression of TNFAIP3 mRNA was increased in A20ZF7 human T cells when compared with paired isogenic control T cells (Figure 6J). The NF-κB family members c-Rel and Rela/p65 bind RORγt promoters and drive RORγt transcription (48). Consistent with increased NF-κB signaling and with our findings in murine Th17 cells, ablation of A20ZF7 in human Th17 cells led to higher expression of RORγt (Figure 6I). Finally, A20ZF7-deleted human Th17 cells led to elevated expression of IL17A and IL22 transcripts relative to paired, isogenic, A20-competent control cells (Figure 6J). Hence, A20’s ZF7 motif restrains RORγt expression and IL22 expression in human Th17 cells.
-
Discussion
Our studies uncover a new spontaneous model of proximal enteritis that links A20’s M1-Ub–binding motif to pathogenic Th17 cell activation, IL-22–dependent epithelial dysfunction, microbe-dependent enteritis, and epigenetic regulation of Il22 transcription. Inflammation of the proximal duodenum in A20ZF7 mice aligns with two human diseases that can afflict this portion of the intestine: Crohn’s disease and celiac disease. Both of these diseases are genetically linked to extragenic polymorphisms upstream of TNFAIP3 (7, 9). While the mechanisms by which A20 regulates intestinal immunity remain incompletely understood, we show here that non-enzymatic Ub binding by A20 ZF7 is more important than A20 ZF4’s Ub binding or E3 Ub ligase activity for preserving small intestinal homeostasis. The selective importance of A20 ZF7’s Ub-binding motif may be related to its preferential binding to M1-linked Ub dimers (23), its increased binding affinity to Ub tetramers (22), and/or its ability to bind and regulate IKKγ signaling complexes (21). While the exact Tnfaip3 mutation we have engineered in A20’s ZF7 motif has not been described in human patients, haploinsufficient HA20 patients harbor many TNFAIP3 mutations that cause premature stop codons that commonly result in the loss of A20’s C-terminal ZF7 domain (10, 47, 49). Therefore, our findings not only highlight the importance of the non-enzymatic ZF7 functions of the A20 protein in intestinal immune homeostasis but also identify a unifying molecular dysfunction that may regulate intestinal disease in human patients.
We have obtained new insights into the homeostasis of intestinal tissue-resident Th17 cells. Expansion of Th17 cells in A20ZF7 mouse intestines may reflect exaggerated TCR responses and increased IL-1β expression. Aberrant TCR and cytokine responses likely drive Th17 cells toward proinflammatory states characterized by the expression of IL-17F, IL-22, GM-CSF, M-CSF, and/or granzymes (50, 51). This important transition can be stimulated by IL-23 (50, 52). The importance of Th17-derived cytokines other than IL-17A in the intestine has been implicated by the clinical antiinflammatory efficacy of IL-23 blockade contrasted with proinflammatory consequences of IL-17A inhibition (53). Our studies highlight the ability of IL-22 to drive proximal enteritis. Increased expression of other Th17 cytokines in A20ZF7 mouse intestines, e.g., IL-17F and/or IFN-γ, may also contribute to enteritis in these mice. More broadly, increased cytokine expression by intestinal A20ZF7 T cells implicates A20ZF7 as a critical mediator of Th17 cell quiescence.
In addition to supporting epithelial cell proliferation and repair, our studies show that IL-22 can also disrupt IEC expression of tight junction proteins and alarmins. Among cytokines that directly regulate IEC functions, IL-22 is distinct from IL-17A, IL-17F, and IFN-γ in that IL-22 binds to IECs and not immune cells (28). In this regard, IL-22 occupies a unique niche in immune-epithelial crosstalk. While prior studies showed that IL-22 supports reparative functions in IECs (2, 54, 55), IL-22 can also mediate inflammation when Treg or IL-10 deficiency causes aberrant macrophage activation and colitis (56). By contrast, A20ZF7 mice develop enteritis in the presence of supranormal levels of Tregs and IL-10. Hence, our studies reveal that IL-22 can cause intestinal inflammation despite intact Treg functions. IL-22–dependent pathophysiology in A20ZF7 mice also appears distinct from that seen in Treg- and IL-10–deficient mice in that A20ZF7 mice develop proximal enteritis, while the latter models predominantly develop colitis. We have found that IL-22 stimulates exaggerated IEC elaboration of alarmins, hyperproliferation of crypt IECs, and compromised epithelial permeability, leading to microbe-dependent enteritis. These epithelial dysfunctions broaden IL-22–dependent biology beyond its tissue-reparative and defensin activities. They demonstrate that dysregulated IL-22 expression from pathogenically activated Th17 cells can profoundly disrupt IEC homeostasis. The context-specific variables that influence whether IL-22 performs proinflammatory versus tissue-reparative functions may include the cellular source of IL-22 (i.e., Th17 versus ILC3 cells), the epithelial subtypes responding to IL-22 (e.g., small versus large intestine, crypt progenitor versus mature epithelial cells), coexpressed cytokines, and/or the abundance or duration of IL-22 signals.
We have found that enteritis in A20ZF7 mice is microbe dependent. Yet this enteritis selectively afflicts the proximal small intestine that is typically colonized with fewer numbers of bacteria than more distal regions. Our studies with neocolonized, or “conventionalized,” A20ZF7 mice most directly establish the microbe-dependence of our enteritis model and provide a platform for future dissection of microbe-driven responses in these mice. Segmented filamentous bacteria (SFB) selectively expand Th17 cells in ilea of mice, and this selectivity is related to this organism’s preferential colonization of this segment of the intestine (37–40). Indeed, we detected SFB in the ilea — but not the duodena — of a few WT and A20ZF7 mice. Hence, it is more likely that microbes that selectively colonize the proximal small intestine may drive duodenitis in A20ZF7 mice. Small-intestinal microbes have recently been highlighted to alter lipid absorption (57). Alternatively, IECs in the proximal small intestine of these mice may harbor preferential sensitivity to microbially triggered IL-22 signals. Other potential contributing factors include luminal products that are concentrated in the proximal bowel, e.g., bile acids emptied into the duodenum via the bile duct. As we have found that germ-free conditions abrogate enteritis, these moieties could be secondary bile acids that are modified by small intestinal bacteria. Future studies of duodenal microbes and/or bile acid metabolites could unveil mechanisms by which homeostasis is preserved in the duodenum.
We have uncovered T cell–autonomous functions of A20 ZF7 that restrain pathogenic expression of IL-22. We have observed increased STAT3 phosphorylation in in vitro–differentiated A20ZF7 Th17 cells, suggesting that A20ZF7 may restrain IL-6/IL-21–induced JAK/STAT signals in these cells. Prior work from our laboratory and others showed that A20 restrains TCR-induced NF-κB signaling in naive T cells (15, 18, 58). Hence, intestinal A20ZF7 Th17 cells may exhibit enhanced NF-κB signaling in response to homeostatic TCR signals. Notably, the enhanced Il22 transcription in in vitro–differentiated A20ZF7 Th17 cells occurs in the absence of IL-1 or IL-23, implicating A20ZF7 in the restraint of Th17 cell responses independent of these pathogenic cytokines. Prior studies showed that components of NF-κB — c-Rel and Rela/p65 — directly promote the expression of RORγt (48). Our current results are consistent with the idea that increased NF-κB activity in A20ZF7 Th17 cells stimulates increased RORγt expression. Our data further indicate that increased nuclear RORγt directly promotes Il22 enhancer activity that enhances Il22 transcription in A20ZF7 T cells. This enhancer has been described to harbor NF-κB, RORγt, Runx1, and AP-1 binding sites (44). Intriguingly, this murine enhancer is also highly conserved upstream of the human IL22 locus. Combined with our finding that ablation of A20ZF7 in human T cells causes increased IL22 expression, inhibition of this enhancer by A20ZF7 prevents proinflammatory expression of IL22 and may restrain a program of pathogenic activation of both murine and human Th17 cells Given the importance of pathogenically activated Th17 cells to intestinal inflammation, this biochemical function provides an important new lever for preserving Th17 cell quiescence and preventing human disease.
-
Methods
Sex as a biological variable. Both male and female mice were used in this study, and similar results were obtained with both. Sex was not considered a biological variable in this study.
Mice. Animal studies were conducted under an approved Institutional Animal Care and Use Committee protocol at UCSF. Mice were bred and housed at 22°C under a 12-hour light/12-hour dark cycle with ad libitum access to food and water. A20C103/C103 (OTU), A20ZF4/ZF4 (ZF4), and A20ZF7/ZF7 (ZF7) knockin mice were generated in our laboratory and previously described (20, 21). B6.129S2 Ighmtm1Cgn/J (μMT; 002288), B6.129S7 RAG-1tm1Mom/J (Rag1; 002216), and C57BL/6 Il22tm1.1(icre)Stck/J (IL22; 027524) mice were purchased from The Jackson Laboratory. Il17a–/– mice were provided by Y. Iwakura (Tokyo University of Science, Tokyo, Japan) via S. Gaffen, University of Pittsburgh, Pittsburgh, Pennsylvania, USA).
Tissue histology and scoring. Mice at the indicated ages were euthanized, and the proximal small intestine was collected in 4% paraformaldehyde. Samples were subsequently processed and stained by HistoWiz to produce H&E-, CD4-, or Ki67-stained slides. Pathology scores of intestinal inflammation were generated by assessment of total tissue inflammation and epithelial changes in the colon. The total inflammation score (scale 0–3) was based on the overall severity or extent of inflammation in the intestine. An epithelial change score (scale 0–4) was assigned for each of the following categories: goblet cell loss, intraepithelial neutrophils and/or cryptitis, abscesses, and crypt loss. The scores were summed to generate a pathology score for each mouse.
Radiation bone marrow chimeras. At 6 weeks of age, WT recipient mice were irradiated with 12 Gy total-body radiation. The same day, recipients were injected intravenously with 5 × 106 bone marrow cells isolated from femora and tibiae of corresponding donors (WT or A20ZF7 mice). Animals were kept on antibiotics in the drinking water for 1 week after irradiation. Bone marrow recipients were sacrificed 12 weeks after bone marrow transfer.
FITC-dextran epithelial barrier assay. Mice (11 weeks old) were fasted for 5 hours before oral gavage of FITC-dextran (average MW 4,000; Sigma-Aldrich 46944) at 500 mg/kg body weight. Serum was collected 4 hours after gavage, and the fluorescence intensity was measured on a Molecular Devices fluorescence microplate reader.
Isolation of lamina propria cells. Single lamina propria cells were isolated from small intestinal lamina propria as previously described (59). Briefly, the proximal 10 cm of small intestines from euthanized mice were flushed with cold PBS to clear feces and mucus. Excess mesenteric fat and Peyer patches were removed. Intestines were opened longitudinally and incubated in pre-digestion buffer (calcium- and magnesium-free HBSS, 3% FBS, 10 mM HEPES [pH 7.4], 1 mM dithiothreitol, 5 mM EDTA) twice for 15 minutes each, rinse buffer (calcium- and magnesium-free HBSS, 3% FBS, 10 mM HEPES [pH 7.4]) once for 5 minutes, and digestion buffer (RPMI with 3% FBS, 10 mM HEPES [pH 7.4], 0.03 mg/mL DNase I, 0.1 mg/mL Liberase TM (Sigma 5401119001) for 10 minutes, all at 37°C with agitation at 220 rpm. Enzyme-digested tissue was further dissociated in a gentleMACS C-tube using the gentleMACS (Miltenyi Biotec) Dissociator m_intestine program, added to RPMI with 10 mM HEPES (pH 7.4) and 3% FBS, and filtered through 70- or 100-μm filters. Leukocytes were enriched from the interface of a 40%/80% Percoll (GE Healthcare 17-081-01) gradient after centrifugation.
Flow cytometry. Single-cell suspensions were twice rinsed with PBS, stained with an amine-reactive viability dye in PBS for 15 minutes, quenched, and washed with FACS wash buffer (FWB: HBSS or PBS with 0.5% BSA). Cells were then blocked with 2 μg FcBlock (per 1 million cells) for 15 minutes and stained with antibodies against cell surface antigens for 30 minutes. For nuclear assays, nuclei were isolated as previously described (60) with slight modifications: cells were incubated in ice-cold isolation buffer (375 mM sucrose, 10 mM HEPES [pH 7.9], 10 mM potassium chloride, 5 mM magnesium chloride, 0.1% vol/vol Triton X-100, protease and phosphatase inhibitors) on ice for 15 minutes, and nuclei were collected by centrifugation at 1,300g for 5 minutes at 4°C. For intracellular antigens, cells/nuclei were subsequently fixed in 5% neutral-buffered formalin for 30 minutes at room temperature, permeabilized with eBioscience Perm Buffer (Thermo Fisher Scientific 00-8333-56), blocked with normal rat serum (STEMCELL Technologies) for 15 minutes, and rocked with primary antibodies overnight at 4°C. Single cells/nuclei were washed twice with FWB before analysis on a flow cytometer.
The following antibodies were used for flow cytometry: anti-CD45 (30-F11, BD Biosciences), anti-CD90.2 (30-H12, BD Biosciences), anti-TCRβ (H57-597, BioLegend), and anti-CD4 (GK1.5, BioLegend) for staining of mouse cell surface proteins; and anti–IL-17A (TC11-8H4, BioLegend), anti–IL-22 (poly1564, BioLegend), anti-RORγt (B2D, Thermo Scientific; or REA278, Miltenyi Biotec), and anti–phosphorylated Stat3 (4/P-STAT3, BD Biosciences) for staining of human or mouse intracellular proteins. For intracellular cytokine staining, cells were first incubated for 4 hours in medium with 100 ng/mL phorbol 12-myristate 13-acetate (PMA), 1 μg/mL ionomycin, and 5 μg/mL brefeldin A.
Murine in vitro Th17 differentiation. Naive CD4+ T cells were isolated from murine splenocytes using a Mouse CD4 naive T cell negative selection kit (STEMCELL Technologies 19765) per the manufacturer’s instructions. Isolated cells were subsequently differentiated in plates precoated with 2 μg/mL anti-CD3 (overnight at 4°C, or 2 hours at 37°C) in Th17 medium (IMDM with 10% FBS, 50 μM β-mercaptoethanol, 2 μg/mL anti-CD28 [Bio X Cell BE0015, clone 37.51], 20 ng/mL recombinant murine IL-6 [R&D Systems 406-ML], 5 ng/mL recombinant human TGF-β1 [PeproTech 100-21], 10 μg/mL anti–IL-4 [Bio X Cell BE0045, clone 11B11], and 10 μg/mL anti–IFN-γ [Bio X Cell BE0055, clone XMG1.2]) at a density of 1 million cells/mL. Medium was replenished on day 2, and cells were split into fresh medium on day 3 of differentiation to maintain a cell density of 1–2 million cells/mL. For RORγt inhibition experiments, naive CD4+ T cells were differentiated for 6 days before incubation with 1 μM GSK805 (MedChemExpress HY-12776) in fresh Th17 medium for 16 hours prior to harvesting.
RNA isolation and quantitative real-time PCR. Total RNA was isolated from whole intestinal tissue or single-cell suspensions (epithelial fractions or lamina propria) using QIAGEN RNeasy extraction kits or TRIzol (Invitrogen) per the respective manufacturer’s instructions. Whole tissues were lysed in either ice-cold TRIzol or QIAGEN RLT Buffer using Lysing Matrix D (MP Biomedical 116913050-CF) with MP Biomedical FastPrep-24 Homogenizer (1 cycle of 20 seconds at 4 m/s). Single-cell suspensions were lysed directly in TRIzol. RNA was reverse-transcribed using High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific 4368814) per the manufacturer’s instructions and assayed by quantitative real-time PCR using TaqMan probes (Thermo Scientific) or the SYBR Green (Thermo Scientific) method. Relative mRNA expression was calculated using the Livak (ΔΔCt) method, with Actb as a loading control, unless otherwise noted. In experiments using human cells, the housekeeping genes ACTB, GAPDH, EIF4G2, and RPLP0 were collectively used, and the geometric mean was used in calculating relative mRNA expression. To detect segmented filamentous bacteria (SFB), 16S ribosomal RNA (rRNA) was amplified using primer sequences specific to SFB 16S rRNA: 5′-GATCCTGGCTCAGGACGAAC-3′ and 5′-TTCATCGGGCTATCCCCCA-3′. PCR products from ileal samples with detectable levels of SFB 16S rRNA were subjected to agarose electrophoresis, and the 146-bp products were excised, purified using QIAGEN Gel Extraction kit, and Sanger-sequenced. NCBI BLAST of Sanger sequences confirmed the PCR to specifically detect SFB 16S rRNA. 16S rRNA copies were calculated based on a standard curve generated using a double-stranded DNA gBlock template (Integrated DNA Technologies) encoding a portion of the SFB 16S rRNA sequence. The limit of detection of this assay is routinely less than 1 copy of SFB 16S rRNA.
Single-cell RNA sequencing and data analysis. Lamina propria cells were isolated from small intestines of 2 mice of each genotype as described above. Isolated cells were negatively selected for T cells and ILCs (STEMCELL Technologies 19851) and stained with TotalSeq-C anti-mouse hashtagged antibodies (BioLegend). Cells from each animal were separately hashtagged. For each genotype, a total of 60,000 cells were collected for processing using the 10x Genomics Chromium Next GEM Single Cell 5′ Kit v1.1. Sequences were aligned and processed with Cell Ranger v7.1 using the mm10 reference genome and default parameters. Cell Ranger output was further processed with R version 4.3.3 and Seurat version 4.3 (49). Seurat objects were created using only genes that appeared in at least 3 cells. Cells were further filtered to exclude multiplets (defined as having 2 or more different hashtags) and low-quality/multiplet cells (cells with fewer than 200 detected genes, more than 2,500 detected genes, or more than 15% mitochondrial reads). Read counts were then normalized using NormalizeData. After viewing of UMAP clusters, T and innate lymphocyte clusters were selected based on the presence of T/innate lymphocyte genes (Ptprc, Trac, Trdc, Cd3e, Cd4, Cd8a, Ncr1, Klrb1b) as well as the absence of non-T/non-innate lymphocyte genes (Des, Acta2, Col1a2, Pecam1, Cdh5, Epcam, Cd79a, Mcpt1, Apoe).
Cell clusters were generated using the Louvain algorithm implemented by the FindClusters function. Marker genes for each cluster were determined using Wilcoxon’s test on the raw counts, implemented by the function FindAllMarkers, and clusters of cell types were additionally determined by manual inspection of the lists of cluster marker genes. Dimensionality reduction by UMAP was performed using the RunUMAP function with the 30 largest principal components. Visualization of all scRNA-Seq data was generated using the Seurat package and/or ggplot2. To remove cell cycle phases as a source of clustering heterogeneity, Seurat’s CellCycleScoring function was used to score each cell and regress out the effects of cell cycle genes/phases.
Bulk RNA-Seq library preparation and analyses. Total RNA was isolated from intact proximal small intestine as described above. One microgram of total RNA was depleted of rRNA (NEBNext rRNA Depletion Kit v2, New England Biolabs E7405) and subsequently fragmented, reverse-transcribed into cDNA, and amplified into barcoded libraries with NEBNext RNA Library Prep (New England Biolabs E7765) using custom barcoded Illumina-compatible primers. Libraries were pooled and sequenced on an Illumina NovaSeq X Plus instrument as 150-bp paired-end reads.
Sequenced reads (40 million to 70 million paired reads per sample) were processed with fastp (61) for quality control and adapter sequence trimming. Trimmed reads were aligned to the Mus musculus mm10 genome using HISAT2 (62), and transcriptomes for each sample were assembled with StringTie2 (63) and UCSC’s mm10 genes.gtf annotation. After construction of non-redundant transcriptomes across all samples (stringtie --merge), expression counts were tabulated (stringtie -e) and used as input for downstream gene expression analyses. DESeq2 (64) was used for differential gene expression analyses.
Chromatin immunoprecipitation. In vitro–differentiated Th17 cells were processed for chromatin immunoprecipitation (ChIP) as previously described (65). Briefly, 10 million Th17 cells were cross-linked at room temperature in 1% formaldehyde for 8 minutes, quenched with 125 mM glycine for 5 minutes, and resuspended in ChIP lysis buffer (10 mM Tris [pH 8], 1 mM EDTA, 0.5 mM EGTA, 0.5% wt/vol N-lauroyl sarcosine, and protease and phosphatase inhibitors). Each 250- to 300-μL aliquot was individually sonicated in a Bioruptor Pico to generate an average DNA fragment length of 250 bp as determined by agarose electrophoresis. Debris was spun down at 20,000g for 15 minutes, and the cleared chromatin supernatants were quantified and used for subsequent ChIP experiments. Sonicated chromatin was diluted to 500 μL in ChIP lysis buffer with 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM EDTA, and protease and phosphatase inhibitors. Chromatin was precleared with 10 μL protein G Dynabeads (Thermo Fisher Scientific 10003D) for 4 hours at 4°C before incubation with 1 μg of primary antibodies overnight at 4°C. For anti–histone H3 (Abcam ab1791) or anti–acetyl histone H3 lysine 27 (anti-H3K27ac; Active Motif 39133) ChIP, 3 or 10 μg of total chromatin, respectively, was used. Protein G Dynabeads were incubated with immune complexes for 2 hours at 4°C before magnetic capture. Immunoprecipitates were washed 5 times with 1 mL RIPA wash buffer (50 mM HEPES [pH 7.6], 10 mM EDTA, 0.7% wt/vol sodium deoxycholate, 1% vol/vol IGEPAL CA-630 (Sigma-Aldrich), 0.5 M lithium chloride, and protease inhibitors) and once with TE (10 mM Tris with 1 mM EDTA [pH 8]) with 50 mM sodium chloride and eluted in elution buffer (10 mM Tris, 1 mM EDTA, 1% wt/vol sodium dodecyl sulfate, pH 8) at 56°C for 15 minutes. To reverse cross-links, eluates were incubated overnight at 65°C. Samples were diluted 2-fold in TE and sequentially digested with 80 μg of DNase-free RNase A at 37°C for 1 hour, and then 80 μg proteinase K at 56°C for 1 hour. ChIP DNA was purified by phenol/chloroform/isoamyl alcohol (25:24:1) and ethanol precipitation. DNA pellets were resuspended in 10 mM Tris (pH 8) and assayed by quantitative real-time PCR using the SYBR Green method. Antibodies were titrated to ensure they were not limiting.
ATAC-seq library preparation and data analysis. In vitro–differentiated Th17 cells were used to generate Tn5-tagmented DNA fragments as previously described (66). Briefly, to isolate nuclei, cells were lysed in ATAC lysis buffer (10 mM Tris-HCl [pH 7.5], 10 mM sodium chloride, 3 mM magnesium chloride, 0.1% vol/vol IGEPAL CA-630, 0.1% vol/vol Tween-20, 0.01% wt/vol digitonin) for 3 minutes on ice. The nuclei were briefly rinsed in wash buffer (10 mM Tris-HCl [pH 7.5], 10 mM sodium chloride, 3 mM magnesium chloride, 0.1% vol/vol Tween-20) before incubation in transposition buffer (10 mM Tris-HCl [pH 7.6], 5 mM magnesium chloride, 10% vol/vol dimethyl formamide, 0.1% vol/vol Tween-20, 0.01% wt/vol digitonin) with 100 nM loaded hyperactive Tn5 transposase (Diagenode C01070012) for 30 minutes at 37°C at 1,000 rpm. Transposed DNA was isolated from QIAGEN MinElute columns. Libraries were generated using published Illumina-compatible indexed oligonucleotides and 8 cycles of PCR amplification using NEBNext High-Fidelity 2X PCR Master Mix (New England Biolabs M0541). Libraries were purified from SPRIselect beads (Beckman Coulter B23317), and 51-bp paired-end sequences were sequenced on a NovaSeq X instrument. Sequences were aligned to the GRCm38/mm10 mouse reference genome using Bowtie 2 v2.4.5 (67), and PCR duplicates were removed by SAMtools v1.15 (68) followed by Picard v2.27.1 MarkDuplicates (https://broadinstitute.github.io/picard/). BigWig files were generated using UCSC’s bedGraphToBigWig script. DNA accessible regions and sites of differential accessibility were determined by MACS2 v2.2.7.1 (69) (https://pypi.org/project/MACS2/).
CRISPR/Cas9 editing of human CD4+ T cells and in vitro Th17 differentiation. Naive CD4+ T cells were isolated from healthy donor PBMCs using a human CD4 Naive T Cell negative selection kit (BioLegend 480041) per the manufacturer’s instructions. Isolated cells were stimulated in stimulation medium (Immunocult XF serum-free medium plus 50 μM β-mercaptoethanol, Immunocult anti-CD3/-CD28 cocktail [STEMCELL Technologies], and 50 IU/mL human recombinant IL-2 [NIH Biological Resources Branch. Preclinical Biologics Repository]) for 2–3 days. Stimulated cells were subsequently electroporated in a total of 20 μL P3 solution (Lonza) with Cas9 ribonucleoprotein complexes (3.1 μM TruCut Cas9 v2 [Thermo Fisher Scientific] with 9 μM CRISPR guides [Integrated DNA Technologies]) using the Lonza Amaxa 4D-Nucleofector and program EH-115. Target sequences recognized by CRISPR guides were GATACGTCGGTACCGGACCG for the control/non-targeting guide and TTTGGCAATGCCAAGTGCAA and ACCCCCCCAAGCAGCGTTGC for A20ZF7 ablation. Electroporated cells were immediately placed into pre-warmed stimulation medium and incubated for 3 days. Genomic editing was confirmed to be at least 80% efficient by Sanger sequencing. Cells were subsequently differentiated into Th17 cells in plates precoated with 5 μg/mL anti-CD3 (Bio X Cell BE0001-2, clone OKT3) in Th17 medium (Immunocult XF serum-free medium with 50 μM β-mercaptoethanol, 30 ng/mL recombinant human IL-6 [Proteintech HZ-1019], 2.5 ng/mL recombinant human TGF-β1 [PeproTech 100-21], 10 ng/mL recombinant human IL-1β [Proteintech HZ-1164], 10 ng/mL recombinant human IL-23 [Proteintech HZ-1254], 10 μg/mL anti–IL-4 [Bio X Cell BE0240, clone MP4-25D2], and 10 μg/mL anti–IFN-γ [Bio X Cell BE0235, clone B133.5]) at a density of 1 million cells/mL. Cells were split and medium replenished on days 3, 6, and 8 of Th17 differentiation to maintain a cell density of 1–2 million cells/mL. Cells were harvested on day 9 of differentiation for analysis.
Statistics. Statistical analyses were performed using Prism 10 (GraphPad Software). Pathology scores were assessed by an unpaired Mann-Whitney U test. Quantitative real-time PCRs were assessed by a 2-tailed unpaired Student’s t test with Welch’s correction, unless otherwise noted. A significance level of 0.05 was used as the threshold for statistical significance. All data shown are representative of at least 2 independent experiments.
Study approval. All animal studies were conducted under an approved Institutional Animal Care and Use Committee protocol at the University of California, San Francisco (UCSF).
Data availability. Raw bulk RNA-Seq, scRNA-Seq, and ATAC-seq datasets generated for this study were deposited under Gene Expression Omnibus primary accession numbers GSE296200, GSE296201, and GSE296203. The values displayed as individual data points within the quantitative graphs are available in the Supporting Data Values file.
-
Author contributions
CJB, DMS, XS, and HS designed and conducted experiments. EFY, NL, YS, and RA provided technical assistance and conducted experiments. CJB, MCK, and CJY performed and interpreted transcriptomic and epigenetic data. BR contributed mice and helpful discussions. JAT and PJT conducted experiments with germ-free and conventionalized mice. CJB, BAM, and AM conceptualized the overall project, acquired funding, supervised experiments, and wrote and edited the manuscript. The order of co–first authors was determined by the volume and conceptual novelty of the work each contributed.
-
Acknowledgments
We thank members of the Ma laboratory for helpful discussions and advice. We thank Eugene Oltz for helpful discussions. We thank Claire O’Leary for advice regarding isolation of intestinal lamina propria cells. This work was supported by NIH R01CA266755 and the Crohn’s and Colitis Foundation (to AM). Research was supported by equipment available in UCSF’s Parnassus Center for Advanced Technologies. Flow cytometry was performed in UCSF’s Parnassus Flow Cytometry Core (RRID: SCR_018206), which is supported in part by NIH P30DK063720 and NIH S10 Instrumentation Grants S10 1S10OD026940-01 and S10 1S10OD021822-01. Sequencing was performed at UCSF’s Center for Advanced Technologies, which is supported by UCSF Program in Breakthrough Biomedical Research, Research Resource Program Institutional Matching Instrumentation Award, and NIH 1S10OD028511-01 grants.
Address correspondence to: Averil Ma, UCSF, 513 Parnassus Avenue, S-1032C, San Francisco, California 94143, USA. Phone: 415.502.9405; Email: Averil.ma@ucsf.edu.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2025, Bowman et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2025;135(17):e187499. https://doi.org/10.1172/JCI187499.
-
References
- McGeachy MJ, et al. The IL-17 family of cytokines in health and disease. Immunity. 2019;50(4):892–906.
- Keir ME, et al. The role of IL-22 in intestinal health and disease. J Exp Med. 2020;217(3):e20192195.
- Ueno A, et al. Th17 plasticity and its relevance to inflammatory bowel disease. J Autoimmun. 2018;87:38–49.
- Chen L, et al. The role of Th17 cells in inflammatory bowel disease and the research progress. Front Immunol. 2022;13:1055914.
- O’Connor W, et al. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat Immunol. 2009;10(6):603–609.
- Fauny M, et al. Paradoxical gastrointestinal effects of interleukin-17 blockers. Ann Rheum Dis. 2020;79(9):1132–1138.
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678.
- Jostins L, et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124.
- Trynka G, et al. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut. 2009;58(8):1078–1083.
- Taniguchi K, et al. Novel TNFAIP3 microdeletion in a girl with infantile-onset inflammatory bowel disease complicated by a severe perianal lesion. Hum Genome Var. 2021;8(1):1.
- Nambu R, et al. A systematic review of monogenic inflammatory bowel disease. Clin Gastroenterol Hepatol. 2022;20(4):e653–e663.
- Zanatta L, et al. Case Report: An early-onset inflammatory colitis due to a variant in TNFAIP3 causing A20 haploinsufficiency. Front Pediatr. 2022;10:1044007.
- Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430(7000):694–699.
- Boone DL, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5(10):1052–1060.
- Düwel M, et al. A20 negatively regulates T cell receptor signaling to NF-κB by cleaving Malt1 ubiquitin chains. J Immunol. 2009;182(12):7718–7728.
- Tavares RM, et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 2010;33(2):181–191.
- Duong BH, et al. A20 restricts ubiquitination of pro-interleukin-1β protein complexes and suppresses NLRP3 inflammasome activity. Immunity. 2015;42(1):55–67.
- Onizawa M, et al. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat Immunol. 2015;16(6):618–627.
- Hitotsumatsu O, et al. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28(3):381–390.
- Lu TT, et al. Dimerization and ubiquitin mediated recruitment of A20, a complex deubiquitinating enzyme. Immunity. 2013;38(5):896–905.
- Razani B, et al. Non-catalytic ubiquitin binding by A20 prevents psoriatic arthritis–like disease and inflammation. Nat Immunol. 2020;21(4):422–433.
- Wertz IE, et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature. 2015;528(7582):370–375.
- Tokunaga F, et al. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO J. 2012;31(19):3856–3870.
- Verhelst K, et al. Linear ubiquitination in NF-κB signaling and inflammation: what we do understand and what we do not. Biochem Pharmacol. 2011;82(9):1057–1065.
- Skaug B, et al. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol Cell. 2011;44(4):559–571.
- Lee EG, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289(5488):2350–2354.
- Garg AV, et al. MCPIP1 endoribonuclease activity negatively regulates interleukin-17-mediated signaling and inflammation. Immunity. 2015;43(3):475–487.
- Pham TAN, et al. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe. 2014;16(4):504–516.
- Bruewer M, et al. Proinflammatory cytokines disrupt epithelial barrier function by apoptosis-independent mechanisms. J Immunol. 2003;171(11):6164–6172.
- Al-Sadi RM, Ma TY. IL-1beta causes an increase in intestinal epithelial tight junction permeability. J Immunol. 2007;178(7):4641–4649.
- Narros-Fernández P, et al. Interleukin-1 family cytokines at the crossroads of microbiome regulation in barrier health and disease. FEBS J. 2024;291(9):1849–1869.
- Kim DY, et al. Epithelial claudin proteins and their role in gastrointestinal diseases. J Pediatr Gastroenterol Nutr. 2019;68(5):611–614.
- Chelakkot C, et al. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp Mol Med. 2018;50(8):1–9.
- Troeger H, et al. Escherichia coli alpha-haemolysin induces focal leaks in colonic epithelium: a novel mechanism of bacterial translocation. Cell Microbiol. 2007;9(10):2530–2540.
- Hering NA, et al. Yersinia enterocolitica induces epithelial barrier dysfunction through regional tight junction changes in colonic HT-29/B6 cell monolayers. Lab Invest. 2011;91(2):310–324.
- Bücker R, et al. Arcobacter butzleri induces barrier dysfunction in intestinal HT-29/B6 cells. J Infect Dis. 2009;200(5):756–764.
- Klaasen HL, et al. Intestinal, segmented, filamentous bacteria. FEMS Microbiol Rev. 1992;8(3–4):165–180.
- Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498.
- Farkas AM, et al. Induction of Th17 cells by segmented filamentous bacteria in the murine intestine. J Immunol Methods. 2015;421:104–111.
- Sano T, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell. 2015;163(2):381–393.
- Ivanov II, et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126(6):1121–1133.
- Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453(7191):106–109.
- Veldhoen M, et al. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med. 2009;206(1):43–49.
- Sekimata M, et al. Runx1 and RORγt cooperate to upregulate IL-22 expression in Th cells through its distal enhancer. J Immunol. 2019;202(11):3198–3210.
- Withers DR, et al. Transient inhibition of ROR-γt therapeutically limits intestinal inflammation by reducing TH17 cells and preserving group 3 innate lymphoid cells. Nat Med. 2016;22(3):319–323.
- Xiao S, et al. Small-molecule RORγt antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 2014;40(4):477–489.
- Zheng C, et al. Infantile onset intractable inflammatory bowel disease due to novel heterozygous mutations in TNFAIP3 (A20). Inflamm Bowel Dis. 2018;24(12):2613–2620.
- Ruan Q, et al. The Th17 immune response is controlled by the Rel-RORγ-RORγ T transcriptional axis. J Exp Med. 2011;208(11):2321–2333.
- Karri U, et al. The complexity of being A20: from biological functions to genetic associations. J Clin Immunol. 2024;44(3):76.
- Hue S, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203(11):2473–2483.
- Schnell A, et al. Stem-like intestinal Th17 cells give rise to pathogenic effector T cells during autoimmunity. Cell. 2021;184(26):6281–6298.
- Ahern PP, et al. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 2010;33(2):279–288.
- Moschen AR, et al. IL-12, IL-23 and IL-17 in IBD: immunobiology and therapeutic targeting. Nat Rev Gastroenterol Hepatol. 2019;16(3):185–196.
- Eyerich K, et al. IL-17 and IL-22 in immunity: driving protection and pathology. Eur J Immunol. 2017;47(4):607–614.
- Klotskova HB, et al. The role of interleukin-22 in mammalian intestinal homeostasis: friend and foe. Immun Inflamm Dis. 2024;12(2):e1144.
- Gunasekera DC, et al. The development of colitis in Il10–/– mice is dependent on IL-22. Mucosal Immunol. 2020;13(3):493–506.
- Martinez-Guryn K, et al. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe. 2018;23(4):458–469.
- Giordano M, et al. The tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20) imposes a brake on antitumor activity of CD8 T cells. Proc Natl Acad Sci U S A. 2014;111(30):11115–11120.
- O’Leary CE, et al. Interrogating the small intestine tuft cell-ILC2 circuit using in vivo manipulations. Curr Protoc. 2021;1(3):e77.
- Wysocka J, et al. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol Cell Biol. 2001;21(11):3820–3829.
- Chen S, et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890.
- Kim D, et al. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360.
- Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295.
- Love MI, et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550.
- Bowman CJ, et al. Foxk proteins repress the initiation of starvation-induced atrophy and autophagy programs. Nat Cell Biol. 2014;16(12):1202–1214.
- Corces MR, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14(10):959–962.
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359.
- Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079.
- Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biology. 2008;9(9):R137.
-
Version history
- Version 1 (September 2, 2025): Electronic publication



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)






