Advertisement
Research ArticleDevelopmentVascular biology
Open Access |  10.1172/JCI187532
10.1172/JCI187532
Defective Notch1 signaling in endothelial cells drives pathogenesis in a mouse model of Adams-Oliver syndrome
Alyssa F. Solano,1,2,3 Kristina Preusse,1 Brittany Cain,1 Rebecca Hotz,1 Parthav Gavini,1 Zhenyu Yuan,4 Benjamin Bowen,5 Gabrielle Maco,5 Hope Neal,5 Ellen K. Gagliani,5 Christopher Ahn,6 Hee-Woong Lim,6,7 Laura Southgate,8,9 Rhett A. Kovall,4 Raphael Kopan,1,7 and Brian Gebelein1,7
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Solano, A. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Preusse, K. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Cain, B. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Hotz, R. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Gavini, P. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Yuan, Z. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Bowen, B. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Maco, G. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Neal, H. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Gagliani, E. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Ahn, C. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by
Lim, H.
in:
PubMed
|
Google Scholar
|

1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Southgate, L. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Kovall, R. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Kopan, R. in: PubMed | Google Scholar
1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
2Medical Scientist Training Program, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
3Graduate Program in Molecular and Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, Ohio, USA.
4Department of Molecular and Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
5Department of Chemistry, Xavier University, Cincinnati, Ohio, USA.
6Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
7Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
8School of Health and Medical Sciences, City St George’s, University of London, United Kingdom.
9Department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom.
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
Find articles by Gebelein, B. in: PubMed | Google Scholar
Published October 7, 2025 - More info
J Clin Invest. 2025;135(23):e187532. https://doi.org/10.1172/JCI187532.
© 2025 Solano et al. This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Received: October 8, 2024; Accepted: September 30, 2025
-
Abstract
Adams-Oliver syndrome (AOS) is a rare congenital disorder characterized by scalp, limb, and cardiovascular defects. Although variants in the NOTCH1 receptor, DLL4 ligand, and RBPJ transcription factor have been implicated in AOS, the driving tissue types and molecular mechanisms by which these variants cause pathogenesis are unknown. Here, we used quantitative binding assays to show that AOS-associated RBPJ missense variants compromise DNA binding but not cofactor binding. These findings suggest that AOS-associated RBPJ variants do not function as loss-of-function alleles but instead act as dominant-negative proteins that sequester cofactors from DNA. Consistent with this idea, mice carrying an AOS-associated Rbpj allele develop dominant phenotypes that include increased lethality and cardiovascular defects in a Notch1 heterozygous background, whereas Notch1 and Rbpj compound heterozygous null alleles are well tolerated. To facilitate studies into the tissues driving AOS pathogenesis, we employed conditional genetics to isolate the contribution of the vascular endothelium to the development of AOS-like phenotypes. Importantly, our studies show that expression of the Rbpj AOS allele in endothelial cells is both necessary and sufficient to cause lethality and cardiovascular defects. These data establish that reduced Notch1 signaling in the vasculature is a key driver of pathogenesis in this AOS mouse model.
-
Introduction
Adams-Oliver syndrome (AOS) is a rare congenital condition characterized by aplasia cutis congenita, which is a thinning and/or absence of skin and skull tissue at the top of the head, and transverse terminal limb truncations (1, 2). In addition, patients with AOS frequently present with heart and vascular defects such as atrial and ventricular septal defects, valve anomalies, aortic and pulmonic stenosis, coarctation of the aorta, patent ductus arteriosus, persistent truncus arteriosus, tetralogy of Fallot, cutis marmorata telangiectatica congenita, portal vein agenesis, portal hypertension, esophageal varices, intracranial hemorrhages, and thrombosis (2, 3). A smaller number of patients with AOS have neurological defects such as microcephaly, ventricular dilation, corpus callosum hypoplasia, periventricular lesions, visual deficits, epilepsy, spasticity, and cognitive impairment (2). Approximately 10% have intrauterine growth restriction (2). Hence, AOS features include a complex mixture of symptoms requiring a multidisciplinary approach to clinical management.
Genetic studies have revealed that approximately 40% of patients with AOS inherit variant alleles in 1 of 6 genes: NOTCH1, DLL4, RBPJ, EOGT, DOCK6, and ARHGAP31 (2). AOS cases caused by variants in NOTCH1, DLL4, RBPJ, and ARHGAP31 are autosomal dominant (4–7), whereas EOGT and DOCK6 variants are autosomal recessive (8, 9). Of these genes, 4 encode components of the Notch signaling pathway, including the receptor NOTCH1, the ligand DLL4, the transcription factor RBPJ, and the EGF domain–specific O-linked N-acetylglucosamine transferase EOGT, which posttranslationally modifies Notch proteins (10). The remaining 2 genes encode proteins that regulate small GTPases, with DOCK6 encoding a guanine nucleotide exchange factor and ARHGAP31 encoding a Rho GTPase-activating protein (4, 8). The relationship between the Notch pathway and small GTPase regulators in AOS pathogenesis is unclear. However, patients with Notch pathway variants have a higher prevalence of cardiovascular defects (49% vs. 13%), whereas patients with pathogenic DOCK6 variants have a higher prevalence of brain anomalies (91% vs. 19%) (2). Overall, AOS pathogenesis remains poorly understood, and no disease-modifying therapies are available.
The canonical Notch pathway converts ligand/receptor interactions into changes in gene expression. Signaling is initiated when a ligand (DLL1, DLL3, DLL4, JAG1, or JAG2 in mammals) on a signal-sending cell binds a receptor (NOTCH1, NOTCH2, NOTCH3, or NOTCH4 in mammals) on a signal-receiving cell (10). Force generated during ligand endocytosis induces a receptor conformation change that allows proteolytic cleavage within the NOTCH transmembrane region to release the Notch intracellular domain (NICD) into the cytoplasm (11). NICD then transits to the nucleus, forms a ternary complex with RBPJ and the coactivator MAML, and activates target genes (11, 12). Conversely, RBPJ can also directly bind corepressors that limit Notch target gene transcription (13–16). Thus, Notch signal strength is largely determined by the number of NICD molecules and competing corepressors within a cell (17–19).
Notch signaling is iteratively used throughout development to regulate the morphogenesis of many organs, including the heart (20), vasculature (21), hematopoietic system (22), nervous system (23), and somite-derived organs (24). In fact, clinical studies have implicated aberrant Notch signaling in an array of health disorders that include AOS, aortic valve disease, hypoplastic left heart syndrome, Alagille syndrome, cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL), Hajdu-Cheney syndrome, spondylocostal dysostosis, and cancer (25, 26). How specific defects in the Notch pathway cause this array of disease is an active area of research.
Given the implication of Notch pathway genes in AOS and the observed vascular changes in patients with AOS (2, 27), some have speculated that impaired vascular development drives AOS pathogenesis (6, 28, 29). However, a vascular etiology for AOS has yet to be established, and the heart, skin/scalp, and limb defects found in AOS could be caused by defective Notch signaling in multiple cell types (25, 26). Unfortunately, loss of a Notch1 allele in mice is not sufficient to recapitulate AOS-like phenotypes, whereas loss of a Dll4 allele is so severe that heterozygotes rarely survive to birth due to catastrophic vascular defects (30, 31). Tissue-specific induction of Dll4 heterozygosity within the second heart field has been used to bypass early lethality and model the impact of Dll4 heterozygosity on mouse heart development (32), but the requirement for tissue specificity limits the applications of this model. Thus, we currently lack a good mouse model of AOS to study pathogenesis.
Molecular genetic studies of patients with AOS have revealed frameshift and early truncation defects in NOTCH1 and DLL4 likely to render each allele null (2). These findings are consistent with dominant NOTCH1 and DLL4 variants creating loss-of-function alleles and haploinsufficiency causing AOS (33). In contrast, all AOS-associated RBPJ variants are missense substitutions; no frameshift or nonsense RBPJ variants have been identified that would encode obvious null alleles. To understand the mechanisms by which AOS-associated RBPJ variants affect Notch signaling, we previously leveraged a Drosophila line with an E137V mutation in Suppressor of Hairless [Su(H), fly ortholog of RBPJ] that is analogous to an AOS-associated variant in human RBPJ at residue E63 (5, 34). Intriguingly, a single Su(H)E137V allele was sufficient to induce wing nicking, a phenotype not seen in flies with a single Su(H)-null allele. Moreover, the Su(H)E137V allele dramatically enhanced a loss of sensory bristle phenotype associated with haploinsufficiency of the antagonistic Hairless (H) corepressor, whereas the Su(H)-null allele suppressed this phenotype (34, 35). Molecularly, we found that both the fly Su(H)E137V protein and a mouse RbpjE89G protein that is analogous to the human RBPJE63G AOS variant decreased DNA binding but not NICD nor corepressor binding (34). Consistent with these findings, RbpjE89G did not activate Notch reporter expression as well as WT Rbpj, even though RbpjE89G is properly localized to the nucleus and interacts with full-length NICD1 and the Sharp corepressor as well as WT Rbpj in co-IP assays (34). Taken together, these Drosophila, cell culture, and biochemical findings suggest that RBPJ AOS alleles encode dominant-negative proteins that dysregulate Notch signaling by sequestering NICD and other cofactors from DNA. However, whether cofactor sequestration is consistent across all AOS-associated RBPJ variants and how this mechanism leads to the complex array of AOS symptoms in humans is not understood.
Here, we used quantitative DNA binding assays to show that all 6 AOS-associated RBPJ alleles encode proteins with defective DNA binding activity but with differing degrees of severity, ranging from a 3-fold decrease to complete loss in DNA binding. To assess how such alleles affect mammalian development, we made 2 mouse models that encoded analogous AOS-associated RBPJ variants with approximately 3-fold (RBPJS358R) and approximately 6-fold (RBPJE89G) decreased DNA binding activity. Characterization of these mice revealed that, while each allele compromised the Notch pathway, they were insufficient to cause dominant phenotypes in an otherwise WT background. However, mice that were compound heterozygous for a Notch1-null allele and the RbpjE89G allele had decreased viability and showed pronounced vascular and heart defects. In contrast, compound heterozygous mice with Notch1- and Rbpj-null alleles were born at normal Mendelian ratios and showed no gross morphological defects. These findings are consistent with AOS-associated Rbpj variants encoding dominant-negative proteins and not null alleles. Since an Rbpj-null allele is well tolerated in mice, we used conditional genetics to demonstrate that expressing the RbpjE89G dominant-negative allele in endothelial cells is both necessary and sufficient to induce lethality due to vascular and heart-related defects. These studies provide mechanistic insights into how defective Notch signaling in the endothelium causes pathogenesis in mice and thereby serves as a useful model to study human AOS pathogenesis.
-
Results
AOS-associated RBPJ variants reduce DNA but not cofactor binding. RBPJ has a conserved core consisting of an N-terminal domain (NTD), beta-trefoil domain (BTD), interdomain linker, and C-terminal domain (CTD) (Figure 1A). In the human ortholog (NM_005349.4), residues 57–67 and 165–170 in the NTD and 192–197 in the BTD directly interact with DNA (Figure 1, A and B) (36). To date, 6 likely deleterious RBPJ variants have been reported in AOS, all of which are missense substitutions that alter highly conserved residues (Y60C, E63G, R65G, F66V, K169E, and S332R; Figure 1, A and B) (2, 5). Five of these missense variants occur within the RBPJ DNA binding domain, whereas S332R occurs within the linker region (Figure 1, A and B). Consistent with the locations of these point mutations, prior studies characterized the DNA binding properties of two RBPJ disease variants (E63G and K169E) and found decreased DNA binding (5). These studies led to the prediction that AOS-associated RBPJ variants behave as loss-of-function alleles due to decreased DNA binding.
 Figure 1
Figure 1AOS-associated RBPJ variants impair DNA binding. (A) Domain map and sequence alignment of RBPJ orthologs. Conserved residues are highlighted, and AOS-associated variants (*) are denoted by human (blue) and mouse (orange) residue numbers. Black bars indicate DNA-binding regions. NTD = N-terminal domain. BTD = beta-trefoil domain. CTD = C-terminal domain. (Created in BioRender.) (B) Structure of RBPJ on DNA with AOS-associated residue changes denoted by human (blue) and mouse (orange) numbers. (C–H) PyMOL models of structural changes and representative comparative EMSAs of AOS-associated RBPJ variants. Dashed lines within each model denote DNA-residue or residue-residue polar interactions, and red discs indicate steric clash. EMSAs were performed using equimolar concentrations (5, 25, and 125 nM) of WT mouse RBPJ and the R91G (C), K195E (D), E89G (E), Y86C (F), F92V (G), and S358R (H) variants on a DNA probe encoding a high-affinity RBPJ binding site. (I) Graph quantifying the probe depletion for each variant across triplicate EMSAs (see Supplemental Figure 1). A 1-way ANOVA with Tukey’s post hoc correction was used to compare WT RBPJ with each variant. (J) Tabulated ITC data measuring DNA binding affinity of RBPJ variants. Fold-change calculated relative to WT RBPJ. A 2-tailed t test was used to compare KD of WT RBPJ to each variant. *P < 0.05, **P < 0.01, ****P < 0.0001. NS, not significant. N/A, not applicable.
To determine whether all RBPJ AOS variants affect DNA binding and to directly compare the binding activity of each variant, we performed electrophoretic mobility shift assays (EMSAs) and isothermal titration calorimetry (ITC) assays using DNA probes encoding an RBPJ binding site and purified AOS-associated RBPJ variants within the context of the mouse protein (Figure 1 and Supplemental Figures 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/JCI187532DS1). In addition, we modeled each variant in the context of the known RBPJ/DNA structure to better understand the molecular nature of each defect (Figure 1, C–H). Note, we previously reported ITC assays to assess the DNA binding affinity of WT RBPJ and the RBPJE89G and RBPJK195E AOS variants (34). We included that data here along with new EMSA data for comparative purposes, and we have cited the original source as appropriate. Collectively, these studies revealed 2 findings: first, all variants significantly decreased DNA binding compared with WT RBPJ; and second, the variants’ impact on DNA binding fell across a spectrum of severity (Figure 1, C–J, and Supplemental Figures 1 and 2). Below, we describe the impact of each variant.
The most severe variant was RBPJR91G, which abolished DNA binding in EMSAs (Figure 1, C and I) and ITC assays (Figure 1J and Supplemental Figure 2A). This finding is congruent with the R91G change being predicted to abolish polar interactions with both DNA and the adjacent E89 residue (Figure 1C). Almost as severe was RBPJK195E, which significantly compromised DNA binding in EMSAs (Figure 1, D and I) and decreased binding approximately 16-fold in ITC assays (Figure 1J and Supplemental Figure 2A). Consistent with this dramatic loss in DNA binding, the K195E change introduced electrostatic repulsion and steric clashing within a region involved in direct binding to the DNA backbone (Figure 1D).
The RBPJE89G and RBPJY86C variants decreased DNA binding to a similar extent in EMSAs (Figure 1, E, F, and I). ITC assays further showed that RBPJE89G resulted in an approximately 6-fold loss in DNA binding relative to WT RBPJ (Figure 1J and Supplemental Figure 2A). Consistent with these findings, the E89G change is predicted to abolish polar interactions with Y86 and R91. Unfortunately, we were unable to purify sufficient RBPJY86C to perform ITC assays. Moreover, the RBPJY86C/DNA complex migrated slower than WT RBPJ and all other tested variants in EMSAs, even though these proteins were similar in size in SDS gels (Supplemental Figure 1B). Since RbpjY86C introduces a Cys residue, we treated the protein with reducing agents and performed EMSAs but did not observe a change in this slower migration pattern (Supplemental Figure 1C). Although it is unclear why the Y86C substitution results in a slower migrating band, the similar loss of affinity observed by RBPJY86C and RBPJE89G in EMSAs is consistent with structural analysis showing that Y86C is predicted to disrupt polar and nonpolar interactions with DNA (Figure 1F).
The last 2 variants, RBPJF92V and RBPJS358R, resulted in weaker but still significant decreases in DNA binding in EMSAs compared with WT RBPJ (Figure 1, G and H). ITC assays confirmed an approximately 3-fold decrease in DNA binding affinity for each variant (Figure 1J and Supplemental Figure 2A). The modest impact on DNA binding is consistent with S358R residing in a region that does not directly contact DNA. However, this variant is predicted to induce steric clashing with surrounding residues (Figure 1H) and thereby could cause protein folding changes that result in decreased DNA binding. The F92V variant is not predicted to change polar interactions or introduce steric clashing. However, F92 appears to have substantial nonpolar interactions with the DNA backbone that the smaller V92 residue may not fully recapitulate (Figure 1G). Taken together, these DNA binding assays show that all RBPJ AOS variants negatively affect DNA binding but to varying degrees.
These DNA binding assays support the idea that RBPJ AOS alleles encode defective transcription factors that fail to properly bind DNA. In addition to binding DNA, RBPJ directly recruits NICD to activate transcription and corepressors to inhibit transcription. We previously showed that 2 AOS variants, RBPJE89G and RBPJK195E, do not significantly alter their affinity for the NICD1 coactivator or the SHARP corepressor (34). Here, we found that RBPJF92V binds both NICD1 and SHARP with similar affinities as WT RBPJ and that RBPJR91G binds NICD1 with a similar affinity as WT RBPJ in ITC assays (Supplemental Figure 2, B and C, and Supplemental Table 1). Since Y86C is similarly found far from the NICD and SHARP interaction regions, this variant is also unlikely to alter cofactor binding. However, because S358R is located within a region not directly associated with DNA or cofactor binding, we tested RBPJS358R in ITC assays (Supplemental Figure 2, B and C) and found that it also binds NICD1 and SHARP with the same affinity as WT RBPJ (Supplemental Table 1). Thus, all RBPJ variants associated with AOS negatively affect DNA binding but not cofactor binding, consistent with the model that RBPJ AOS variants act as dominant-negative proteins that sequester cofactors away from WT RBPJ and off DNA.
RbpjE89G and RbpjS358R mouse models reveal that phenotypic severity correlates with loss in DNA binding affinity. To make mouse models with AOS-associated Rbpj alleles, we used CRISPR/Cas9 gene editing to engineer 2 Rbpj mutations. We chose to model the RbpjS358R and RbpjE89G variants based on their mild (approximately 3-fold loss) and moderate (approximately 6-fold loss) impacts on DNA binding affinity, respectively, to avoid potential heterozygote lethality in a mouse carrying a severe variant. To introduce S358R (human S332R), we used a donor sequence to replace part of exon 9 of mouse Rbpj (Figure 2A). We similarly introduced E89G (human E63G) using a donor sequence to replace part of exon 3 (Figure 2B). In both cases, silent mutations were included to introduce restriction enzyme sites that facilitate genotyping, and each variant was confirmed by sequencing (Figure 2, A and B). Note, RbpjS358R was generated on a WT Rbpj allele, and we created RbpjE89G on the well-characterized Rbpjfl allele (37). Our rationale for making RbpjE89G on the floxed allele is that Cre can be used to convert the dominant-negative RbpjE89G,fl allele into an Rbpjnull allele in select tissues of heterozygous mice that still have a non-floxed WT Rbpj allele (i.e., Rbpj+/E89G,fl).
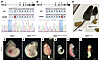 Figure 2
Figure 2Generation of AOS-associated Rbpj variant mouse models reveals impaired animal growth and development. (A) (top) Schematic of mouse Rbpj, detailing the region of exon 9 encoding S358 and the donor sequence used to introduce the S358R substitution. (Created in BioRender.) (bottom) Confirmation of mouse genotype by Sanger sequencing with the codon for S358/R358 highlighted. (B) (top) Schematic of mouse Rbpj, detailing the region of exon 3 encoding E89 and the donor sequence used to introduce the E89G substitution. (Created in BioRender.) (bottom) Confirmation of mouse genotype by Sanger sequencing with the codon for E89/G89 highlighted. (C) Image showing that a typical P17 RbpjS358R/null hemizygote (right) is much smaller than its Rbpj+/+ littermate (left). (D–I) Stereoscope images of E10.5 embryos show that RbpjE89G,fl/E89G,fl homozygotes (F–H) display growth retardation, hemorrhage, pallor, and/or pericardial edema of variable severity. Rbpjnull/null homozygotes (I) show similar, albeit more severe, defects. Scale bar: 1 mm.
To determine the impact of these Rbpj alleles on mouse viability, we assessed offspring for deviation from expected Mendelian ratios. These studies revealed that Rbpj+/S358R heterozygous and RbpjS358R/S358R homozygous mice were viable and occurred at expected ratios (Table 1). Moreover, these mice did not show gross morphological defects, although RbpjS358R/S358R mice were initially smaller than littermates but were of normal size by P5 (Supplemental Figure 3A). We subsequently crossed RbpjS358R/S358R mice with mice carrying an Rbpj-null allele (Rbpj+/null) and found that RbpjS358R/null hemizygotes had significantly reduced viability (Table 1), and surviving offspring were much smaller than littermates (Figure 2C and Supplemental Figure 3B). Thus, the RbpjS358R allele behaves as a weak hypomorph in mice.
We similarly assessed the RbpjE89G,fl allele and found that, although heterozygous mice (Rbpj+/E89G,fl) were viable and lacked gross morphological defects, no RbpjE89G,fl/E89G,fl homozygotes were observed among live offspring (Table 1). To determine when RbpjE89G,fl/E89G,fl homozygotes perish, we performed timed collections at E10.5. Although Rbpj+/E89G,fl embryos resembled WT littermates (Figure 2, D and E), we observed a lower-than-expected frequency of RbpjE89G,fl/E89G,fl embryos (Table 1), and all homozygous embryos were much smaller than their littermates (Figure 2, F–H). Western blot analysis of protein isolated from E10.5 RbpjE89G,fl/E89G,fl and WT embryos revealed that RBPJE89G was expressed at normal levels relative to β-actin (Supplemental Figure 4), consistent with prior studies showing that RBPJE89G had similar stability as WT RBPJ in cell culture (34). Visual analysis of these embryos revealed a range of morphological defects that included hemorrhages (Figure 2F, n = 4/8), pericardial edema (Figure 2, G and H, n = 6/8), pallor (Figure 2G, n = 3/8), and incomplete axial rotation (Figure 2H, n = 3/8). The pericardial edema and incomplete axial rotation are reminiscent of Rbpjnull/null embryos (Figure 2I), although RbpjE89G,fl/E89G,fl embryos fare slightly better than Rbpjnull/null embryos. Lastly, we crossed Rbpj+/E89G,fl mice with Rbpj+/S358R mice and observed a dramatic loss of viability in offspring with both the RbpjS358R and RbpjE89G,fl alleles (RbpjS358R/E89G,fl, Table 1). Altogether, these data show that the RBPJE89G variant, which has an approximately 6-fold decrease in DNA binding activity, causes more severe phenotypes in mice than the RBPJS358R variant with an approximately 3-fold loss in DNA binding.
A compound heterozygous mouse model carrying RbpjE89G and N1null AOS alleles has vascular and heart phenotypes. Our data with the RbpjS358R and RbpjE89G,fl alleles revealed that neither was sufficient to cause dominant AOS-like phenotypes. In contrast, patients heterozygous for analogous RBPJ variants have dominant AOS phenotypes, although the RBPJS332R allele shows incomplete penetrance with only a single symptomatic patient and nonsymptomatic parent (2). These findings are consistent with prior studies showing differences in sensitivity to Notch pathway alleles between mice and humans. For example, NOTCH1 haploinsufficiency can cause human disease such as AOS and aortic valve disease (25), whereas a Notch1-null (N1-null) allele is well tolerated in heterozygous mice (33, 38). Interestingly, a family with AOS was found to have compound heterozygous mutations in both RBPJ and NOTCH1 alleles (2). Hence, we crossed Rbpj+/E89G,fl mice with mice heterozygous for either an N1-null allele that deletes amino acids 1056–2049, thereby removing several EGF repeats, the transmembrane domain, and Ankyrin repeats (N1tm1Con; ref. 38; referred to here as N1null), or an N1-null allele that deletes the promoter and exon 1 (N1tm2Agt; ref. 39; referred to here as N1gKO). Importantly, we observed a dramatic loss of viability in both N1+/null Rbpj+/E89G,fl and N1+/gKO Rbpj+/E89G,fl compound heterozygous mice (Table 2), and the surviving mice generally failed to thrive. Intriguingly, a subset of the N1+/gKO Rbpj+/E89G,fl mice, which had considerable C57/BL6 in their background, had obvious morphological skin/scalp defects (Figure 3, A and B). These findings raise the possibility of genetic background contributing to the skin/scalp defect. Hence, in this study, we focused on identifying the mechanisms of embryonic lethality, which was observed with both N1 alleles in outbred backgrounds.
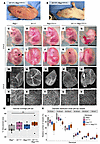 Figure 3
Figure 3N1+/null Rbpj+/E89G,fl embryos display vascular phenotypes. (A and B) Representative images of dorsal midline skin lesions in P0 (A) and P11 (B) N1+/gKO Rbpj+/E89G,fl mice. (C–G) Representative images of E14.5 embryos for WT (Rbpj+/+), N1+/null, and Rbpj+/E89G,fl single heterozygotes and N1+/null Rbpj+/E89G,fl compound heterozygotes. Note, areas of hemorrhage (arrows) are observed in E14.5 N1+/null Rbpj+/E89G,fl embryos but not in control embryos. (H–L) Representative images of E14.5 embryos within their yolk sac for the indicated genotypes. Note, the compound heterozygous embryos have reduced or absent yolk sac vasculature (filled arrowheads). (M–Q) Representative 4× original magnification images of CD31-stained yolk sacs from E10.5 embryos for the indicated genotypes. (R–V) Representative 10× original magnification images of CD31-stained yolk sac microvasculature from E10.5 embryos for indicated genotypes. Scale bars: 0.5 cm (C–L), 1 mm (M–Q), and 100 μm (R–V). (W) Percentage of vascular coverage of yolk sacs measured in representative areas for 5–7 embryos per each indicated genotype. Each dot represents the yolk sac from an individual embryo, and the box plot shows the median with the 25th and 75th quartile highlighted. (X) Distribution of vessels by diameter using representative 400 μm × 400 μm areas of the yolk sac capillary networks stained for CD31. Vessel diameters were assessed between all branch points and measured using the NIS-Elements measurements tool. Each dot represents the yolk sac from an individual embryo, and the box plot shows the median with the 25th and 75th quartile highlighted.
 Table 2
Table 2Impact of Rbpj variants on prenatal and postnatal mouse viability in Notch1-sensitized backgrounds
We next assessed the specificity of the genetic interactions between N1 and RbpjE89G by performing 2 additional tests. First, we crossed each N1-null allele with mice carrying an Rbpj-null allele and found that neither N1+/null Rbpj+/null nor N1+/gKO Rbpj+/null were significantly underrepresented (Table 2). Moreover, unlike the N1 and Rbpj+/E89G,fl compound heterozygotes that showed morphological defects and failed to thrive, the N1+/null Rbpj+/null and N1+/gKO Rbpj+/null compound heterozygous mice were indistinguishable from littermate controls. Thus, the decreased viability observed in the N1 and Rbpj+/E89G,fl compound heterozygotes was due to the presence of the RbpjE89G,fl allele and not simply due to loss of a WT Rbpj allele. Second, we crossed the Rbpj+/E89G,fl allele into a Notch2-sensitized (N2-sensitized) background and observed expected numbers of N2+/lacZ Rbpj+/E89G,fl compound heterozygotes that showed no gross morphological defects (Table 2). Thus, the RbpjE89G allele genetically interacts with N1-null alleles to cause decreased viability but not with an N2-null allele. These data are consistent with clinical findings showing that RBPJ variants cause a NOTCH1-like syndrome (AOS) but not a NOTCH2-like syndrome (Alagille syndrome) (25, 40).
The decreased viability and failure of N1+/null Rbpj+/E89G,fl mice to thrive made it difficult to obtain sufficient mice to perform quantitative analyses of postnatal tissues. To define the cause of lethality in N1+/null Rbpj+/E89G,fl compound heterozygotes, we first genotyped embryos from timed harvests at E10.5, E14.5, and E16.5 to assess the time of embryonic demise. These experiments revealed a gradual decrease in N1+/null Rbpj+/E89G,fl compound heterozygous embryos that became significant by E16.5 (Table 2). Moreover, gross morphological analysis of these embryos revealed vascular phenotypes that included hemorrhages (Figure 3, C–G) and a dramatic reduction in large vessels within the yolk sac vasculature (Figure 3, H–L). Since loss of large vessels could be caused by a lack of vascular remodeling, we stained yolk sacs from E10.5 embryos for the endothelial marker CD31 (Figure 3, M–V). Low magnification images confirmed an overall decrease in large vessels within the yolk sacs of N1+/null Rbpj+/E89G,fl embryos (Figure 3, P and Q) compared with single heterozygous and WT littermates (Figure 3, M–O). However, higher magnification images revealed a robust network of yolk sac capillary vessels in all embryos including N1+/null Rbpj+/E89G,fl compound heterozygotes (Figure 3, R–V). This capillary bed initially forms via vasculogenesis prior to E8.5 and then undergoes N1-dependent remodeling between E8.5 and E10.5 to form a branched hierarchical network of large and small vessels (41). Comparative analysis of the capillary bed revealed that, while the WT and single heterozygous yolk sac vessels had successfully undergone remodeling to form a network of different sized vessels (Figure 3, R–T), the N1+/null Rbpj+/E89G,fl compound heterozygotes showed a range of phenotypes consistent with a lack of or partial failure to undergo hierarchical vascular patterning (Figure 3, U and V, respectively). We next quantified the percentage of vascularized area and the diameter distribution of capillary vessels in the yolk sacs from at least 5 embryos per genotype. Although this analysis revealed that, as a group, the N1+/null Rbpj+/E89G,fl yolk sac capillary bed vasculature was not significantly different from littermate controls (Figure 3, W and X), the N1+/null Rbpj+/E89G,fl embryos showed greater phenotype variability than control embryos. These data are consistent with N1+/null Rbpj+/E89G,fl compound heterozygotes having a partially penetrant disruption or delay in remodeling of the early vascular plexus.
Since heart defects are common in both humans and mice with Notch pathway mutations, we analyzed E16.5 hearts and observed malformations that included ventricular septal defects (VSDs) and dilated coronary vessels in N1+/null Rbpj+/E89G,fl embryos (Figure 4, A–D; we quantify these defects below). We confirmed that the dilated structures in N1+/null Rbpj+/E89G,fl hearts were blood vessels using the endothelial marker VE-cadherin (Fih–I). Consistent with these data, analysis of the hearts from the relatively few P7 N1+/null Rbpj+/E89G,fl mice revealed that one-third also had VSDs (2 of 6, Figure 4, J–N). Although NOTCH1 variants in humans have been associated with bicuspid valve disease, we did not observe obvious valve abnormalities in the hearts of either E16.5 or P7 N1+/null Rbpj+/E89G,fl animals. Altogether, these data demonstrated that N1+/null Rbpj+/E89G,fl mice show increased embryonic lethality that is potentially caused by hemorrhages, diminished yolk sac vascular remodeling, and/or cardiovascular defects.
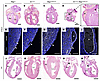 Figure 4
Figure 4N1+/null Rbpj+/E89G,fl embryos display cardiac phenotypes. (A–D) Representative images of E16.5 H&E-stained heart sections from WT (Rbpj+/+), N1+/null, Rbpj+/E89G,fl, and N1+/null Rbpj+/E89G,fl genotypes. The left ventricle (LV) and right ventricle (RV) are labeled, and arrowheads highlight ventricular septal defects in the N1+/null Rbpj+/E89G,fl heart, whereas asterisks highlight dilated coronary vessels. The box in D outlines the region shown at higher magnification at left. (E–I) Representative images of E16.5 heart sections that were stained with VE-cadherin (endothelium, white) and DAPI (nuclei, blue). Arrows indicate coronary vessels, with the lumens of dilated vessels indicated with asterisks. (J–N) Representative images of P7 H&E-stained heart sections from WT (Rbpj+/+), N1+/null, Rbpj+/E89G,fl, and N1+/null Rbpj+/E89G,fl genotypes. The left ventricle (LV) and right ventricle (RV) are labeled, and arrowheads highlight ventricular septal defects in N1+/null Rbpj+/E89G,fl hearts. Scale bars: 0.5 mm (A–D), 100 μm (E–I), and 1 mm (J–N).
Conditional removal of the RbpjE89G,fl allele from only endothelial cells rescues cardiovascular phenotypes. Two pieces of evidence have led to the hypothesis that AOS is largely a vascular disease. First, patients with AOS with NOTCH1, DLL4, and RBPJ variants frequently have cardiovascular defects (2). Second, mouse and zebrafish studies have shown that N1 and DLL4 signaling are critical regulators of vascular development (25, 26). To test this hypothesis, we developed a conditional AOS “rescue” model that uses Tie2-CreYwa to specifically recombine floxed alleles in the developing endothelium (42), which includes the vascular endothelial cells that form the inner lining of blood vessels and the endocardial cells that line the heart. Tie2 is not active in lymphatic endothelial cells, but it is active in hematopoietic stem cells (43). By crossing N1+/null Tie2-Cre+/Ywa mice with Rbpj+/E89G,fl mice, Cre recombination converts the floxed RbpjE89G,fl allele into an Rbpjnull allele in heterozygous endothelial cells and hematopoietic stem cells that still encode a WT Rbpj+ allele (see schematic in Figure 5A). Since N1+/null Rbpj+/null mice occur in expected numbers (Table 2) and do not show overt phenotypes, this mouse model explicitly tests whether expressing the Rbpj+/E89G,fl allele within endothelial cells and hematopoietic stem cells is required (i.e., necessary) to induce morbidity in an N1+/null background (Figure 5A). Consistent with this idea, N1+/null Rbpj+/E89G,fl Tie2-Cre+/Ywa mice had significantly enhanced viability compared with N1+/null Rbpj+/E89G,fl littermates that lack Tie2-Cre (Table 3). Moreover, the N1+/null Rbpj+/E89G,fl Tie2-Cre+/Ywa mice were indistinguishable from control littermates (Supplemental Figure 5), whereas N1+/null Rbpj+/E89G,fl mice without Tie2-Cre generally failed to thrive (Table 3). Thus, Tie2-Cre can significantly rescue the lethality seen in N1+/null Rbpj+/E89G,fl mice by converting the Rbpj+/E89G,fl AOS allele into an Rbpj+/null allele within the endothelium.
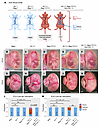 Figure 5
Figure 5Conditional removal of RbpjE89G from the endothelium rescues vascular phenotypes. (A) Schematic of AOS rescue model. Both WT (N1+/+ Rbpj+/+) and N1+/null Rbpj+/null mice are viable and without overt defects. Mice with the N1+/null Rbpj+/E89G,fl genotype have reduced viability, vascular defects, and heart defects (see Table 3 and Figures 3 and 4). A mouse that recombines N1+/null Rbpj+/E89G,fl to N1+/null Rbpj+/null in the endothelium using Tie2-CreYwa tests the necessity of the variant in the vascular endothelium for the development of AOS-like phenotypes. (Created in BioRender.). (B–K) Representative images of E14.5 embryos (B–F) and E16.5 embryos (G–K) within their yolk sac for the indicated genotypes. Note, only the N1+/null Rbpj+/E89G,fl embryos have reduced or absent yolk sac vasculature. The ratio of affected to total individuals is listed in the lower left corner of each panel. (L and M) Visualization of the proportion of embryos with yolk sac vasculature defects at each stage. P values calculated with Fisher’s exact test are noted; NS, not significant.
 Table 3
Table 3Impact of Rbpj variants in the vasculature on prenatal and postnatal mouse viability in Notch-sensitized backgrounds
Because few N1+/null Rbpj+/E89G,fl mice without Tie2-Cre survive postnatally, we quantified the impact of converting the Rbpj+/E89G,fl allele into an Rbpj+/null allele using timed embryo collections at E14.5 and E16.5. Consistent with our postnatal analysis, Tie2-Cre was sufficient to rescue lethality of N1+/null Rbpj+/E89G,fl embryos at E16.5, whereas N1+/null Rbpj+/E89G,fl littermates without Tie2-Cre were significantly underrepresented (Table 3). Moreover, analysis of the yolk sac at both E14.5 and E16.5 revealed that Tie2-Cre significantly rescued the vascular defects of N1+/null Rbpj+/E89G,fl embryos (Figure 5, B–K). For example, although 7 of 9 E14.5 N1+/null Rbpj+/E89G,fl embryos had reduced or absent yolk sac vasculature, 0 of 6 E14.5 N1+/null Rbpj+/E89G,fl Tie2-Cre+/Ywa embryos and none of the control littermates showed diminished yolk sac vasculature (Figure 5, B–F, and L). A similar rescue in yolk sac vasculature was observed in Tie2-Cre positive N1+/null Rbpj+/E89G,fl embryos at E16.5 (Figure 5, G–K, and M). Thus, conditionally converting Rbpj+/E89G,fl into an Rbpj+/null allele with Tie2-Cre was sufficient to rescue both viability and yolk sac vasculature defects in N1+/null heterozygous embryos. Intriguingly, comparative analysis between embryonic time points revealed that the penetrance of yolk sac vasculature defects in the absence of Tie2-Cre was significantly decreased at E16.5 (approximately 33%) compared with E14.5 (approximately 78%) in N1+/null Rbpj+/E89G,fl embryos (P = 0.046). This decreased penetrance in older embryos correlates well with the viability data showing a decrease in the proportion of N1+/null Rbpj+/E89G,fl embryos from E14.5 to E16.5 (Table 2). Hence, these data suggest that those E14.5 embryos with severe yolk sac phenotypes are likely to perish prior to E16.5 and that conditionally deleting the Rbpj+/E89G,fl allele using Tie2-Cre can rescue this phenotype and lethality.
To further assess for possible vascular defects, we immunostained the skin vasculature from the forelimb and scalp regions of E14.5 embryos using a CD31 antibody to label endothelial cells. Analysis of the forelimb tissues for both percentage of vascularized area and branch point density did not reveal significant differences across genotypes (Supplemental Figure 6, A–D). In addition, we analyzed tip cell numbers within the scalp vasculature at E14.5, a time point at which sprouting angiogenesis is actively occurring at the top of the skull, and did not observe any obvious changes in tip cell numbers across genotypes (Supplemental Figure 6, E–I). Thus, although significant defects in the yolk sac vasculature were observed in N1+/null Rbpj+/E89G,fl embryos, we did not observe obvious widespread vascular defects within the embryonic skin.
Next, we assessed whether Tie2-Cre could rescue the heart defects seen in N1+/null Rbpj+/E89G,fl embryos (see Figure 4). Unlike WT embryos (Figure 6A), N1+/null single heterozygotes (Figure 6B), and Rbpj+/E89G,fl single heterozygotes (Figure 6C), N1+/null Rbpj+/E89G,fl compound heterozygotes showed heart defects at E16.5 that included VSDs (5 of 9, Figure 6, D and F) and coronary vessel dilation (5 of 9, Figure 6G). In contrast, we did not observe these phenotypes in N1+/null Rbpj+/E89G,fl Tie2-Cre+/Ywa embryos (Figure 6, E–G), suggesting that the heart and vessel dilation defects in N1+/null Rbpj+/E89G,fl embryos are due to compromised N1 signaling in the developing endothelial and endocardial cells. Together, these results show that expressing the AOS-associated dominant-negative RBPJ protein in the vascular endothelium is necessary to cause cardiovascular phenotypes.
 Figure 6
Figure 6Conditional removal of RbpjE89G from the vascular endothelium rescues heart phenotypes. (A–E) Representative images of E16.5 H&E-stained heart sections. The left ventricle (LV) and right ventricle (RV) are labeled, and an arrowhead highlights a ventricular septal defect in the N1+/null Rbpj+/E89G,fl heart. (F–G) Visualization of the proportion of E16.5 embryos with (F) ventricular septal defects and (G) dilated coronary vessels. P values calculated with Fisher’s exact test are noted. ns = not significant.
Selective induction of N1+/cKO Rbpj+/E89G compound heterozygosity in the vascular endothelium is sufficient to cause lethality and cardiovascular phenotypes. The AOS rescue model reveals that expressing RbpjE89G in the endothelium is necessary to induce morbidity in N1+/null mice. To test whether expressing these alleles within only the endothelium and hematopoietic stem cells is sufficient to induce morbidity, we modified our conditional approach to create an AOS induction model (Figure 7A). First, we used genome editing to remake the RbpjE89G variant on a non-floxed Rbpj allele. Rbpj+/E89G Tie2-Cre+/Ywa mice were then crossed with N1fl/fl mice (44) to generate N1+/flRbpj+/E89G offspring with and without Tie2-Cre. In this model, Tie2-Cre selectively recombines the N1fl/fl allele into a null allele (N1cKO) to induce N1+/cKO Rbpj+/E89G compound heterozygosity within endothelial cells and hematopoietic stem cells of mice that otherwise have 2 copies of N1 (i.e., N1+/fl Rbpj+/E89G) (Figure 7A). Consistent with our hypothesis, N1+/fl Rbpj+/E89G Tie2-Cre+/Ywa mice occurred significantly less often than their littermates, suggesting prenatal demise (Table 4). Moreover, E16.5 N1+/fl Rbpj+/E89G Tie2-Cre+/Ywa embryos had both significantly reduced yolk sac vasculature (Figure 7, B–E) and increased incidences of hemorrhage (Figure 7, F–I) compared with littermates. Additionally, VSDs were observed in N1+/fl Rbpj+/E89G Tie2-Cre+/Ywa hearts but not in control littermates (3 of 7, Figure 7, J–L). Thus, N1+/null Rbpj+/E89G compound heterozygosity in the vascular endothelium is sufficient to cause lethality and cardiovascular defects.
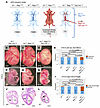 Figure 7
Figure 7Conditional removal of one copy of Notch1 from the vascular endothelium of Rbpj+/E89G mice induces vascular and heart phenotypes. (A) Schematics of AOS induction model. Both WT (N1+/+ Rbpj+/+) and N1+/fl Rbpj+/E89G mice are viable and without overt defects (see Table 4). A mouse that recombines N1+/fl Rbpj+/E89G to N1+/cKO Rbpj+/E89G in the endothelium using Tie2-CreYwa tests the sufficiency of the variant’s presence in the vascular endothelium for the development of AOS-like phenotypes. (Created in BioRender.) (B–E) E16.5 N1+/fl Rbpj+/E89G Tie2-Cre+/Ywa embryos have reduced yolk sac vasculature, increased frequency of hemorrhage (F–I), and ventricular septal defects (J–L). The left ventricle (LV) and right ventricle (RV) are labeled, and an arrowhead highlights a ventricular septal defect in the N1+/fl Rbpj+/E89G Tie2-Cre+/Ywa heart. The ratio of affected individuals to total individuals is listed in the lower left corner of each panel. Scale bars: 0.5 cm (B–D and F–H) and 0.5 mm (J–L). P values calculated with Fisher’s exact test are noted; NS, not significant.
 Table 4
Table 4Impact of Rbpj variants in the vasculature on prenatal and postnatal mouse viability in Notch-sensitized backgrounds
-
Discussion
In this study, we investigated mechanisms underlying how AOS-associated RBPJ variants cause pathogenesis. At the molecular level, we used DNA and protein-protein interaction assays to show that all known AOS-associated RBPJ variants reduce binding to DNA but not to the NICD1 coactivator nor the SHARP corepressor. These in vitro findings are supported by previous co-IP assays showing that full-length NICD1, MAML, and SHARP proteins interact similarly with WT RBPJ and 2 AOS variants (RBPJE89G and RBPJK195E) and that RBPJE89G and RBPJK195E were both properly localized to the nucleus and had similar turnover rates as WT RBPJ (34). At the transcription level, however, titration of a DNA binding–deficient RBPJ variant into cells expressing WT RBPJ lowered Notch-mediated activation, whereas titrating in an RBPJ variant that could neither bind DNA nor NICD1 did not affect transcriptional activation (34). Moreover, a genomic and single-molecule study found that the RBPJK195E AOS variant bound significantly fewer genomic sites and had significantly shorter residency time on DNA than WT RBPJ in HeLa cells (45). Altogether, these biochemical and cellular data support a model whereby AOS-associated RBPJ variants dysregulate Notch signaling by competing for cofactors with WT RBPJ and sequestering them off DNA.
The idea that AOS RBPJ variants act as dominant-negative alleles is further supported by genetic studies. In Drosophila, we previously found that an analogous AOS mutation in the fly RBPJ homologue Su(H) causes dominant Notch phenotypes not observed in flies heterozygous for a Su(H)-null allele (34). Here, we similarly found that mice heterozygous for the RbpjE89G AOS allele suffer lethality and cardiovascular defects in a sensitized N1 background, whereas compound heterozygotes for N1 and an Rbpj-null allele occur in normal ratios and suffer no obvious defects. Lastly, studies of patients with AOS identified 6 missense variants with decreased DNA binding, whereas no mutations have been identified that would render RBPJ into a null allele (2, 5). Moreover, a seventh AOS variant that affects R65 (R65T) was recently reported on ClinVar (VCV001803755.1; https://www.ncbi.nlm.nih.gov/clinvar/), and this variant is likely to negatively affect DNA binding in a manner similar to R65G. Interestingly, however, even though RBPJ-null alleles have not been implicated in AOS, they are underrepresented in the Genome Aggregation Database (pLI = 1; gnomAD v4.1.0) (46). This finding suggests RBPJ haploinsufficiency is likely deleterious in humans, and future studies are needed to determine the impact RBPJ haploinsufficiency has on human development.
Our comparative studies revealed that, while all 6 RBPJ variants compromise DNA binding, they do so to different degrees. These findings predict that RBPJ variants that more strongly decrease DNA binding will result in greater Notch dysregulation and worse outcomes. Consistent with this idea, mice with the RBPJE89G variant that decreases DNA binding 6-fold resulted in more severe phenotypes than mice with the RBPJS358R variant that decreases DNA binding 3-fold. Similarly, the Drosophila Su(H)T4 allele that compromises DNA binding approximately 5-fold resulted in more severe Notch pathway dysregulation compared with the Su(H)O5 allele encoding a protein with approximately 3.5-fold decreased DNA binding (34). Although the rarity of human AOS makes it difficult to perform a comprehensive comparison between variant DNA binding and clinical severity, it is interesting to note that the 2 variants with the weakest impact on DNA binding were found to either have incomplete penetrance (RBPJS332R) or were only found in patients who carried both an RBPJF66V allele and a rare missense N1 allele (2). In contrast, the other RBPJ variants, which impact DNA binding at least 6-fold, have not been associated with other Notch pathway alleles, and to our knowledge all patients with these alleles have AOS phenotypes.
Through conditional genetics, we generated a tractable experimental model ideally suited to identify the defective N1 signaling tissues that contribute to pathogenesis. Our approach takes advantage of the fact that only mice heterozygous for both an N1 and RbpjE89G allele suffer pathological phenotypes. Using Cre recombination, we developed conditional mouse models that either selectively remove the RbpjE89G,fl allele in an otherwise N1+/null background or selectively induce N1+/null Rbpj+/E89G compound heterozygous genotypes in a desired tissue (Figure 5A and Figure 7A). Importantly, Tie2-Cre, which is expressed in endothelial and endocardial cells, rescues lethality and cardiovascular defects by deleting the RbpjE89G,fl allele in an N1 heterozygous background and causes lethality and cardiovascular defects by inducing N1 heterozygosity in the presence of an RbpjE89G allele. While these findings do not preclude the possibility that other cell types contribute to these defects, the fact that having the N1+/null Rbpj+/E89G genotype in the endothelium is both necessary and sufficient to cause AOS-like phenotypes strongly suggests that defective N1-signaling in the vascular endothelium is a major driver of pathogenesis.
These findings raise new questions about what specific cellular processes during vascular and cardiac development are compromised by the RBPJE89G variant. The paucity of large yolk sac vessels in N1+/null Rbpj+/E89G,fl mice suggests a failure to properly remodel the primitive vascular plexus to a hierarchically organized vascular network, a known N1-dependent process (41). In addition, the increase in hemorrhages in these embryos suggests vascular integrity is compromised, similar to that seen with anti-DLL4 antibodies (47) or N1 loss-of-heterozygosity models (48). In contrast, we did not observe obvious defects in sprouting angiogenesis as revealed by tip/stalk cell specification and vascularized branching within skin preparations. However, additional quantitative studies with temporal control using inducible Cre lines are needed to provide a better assessment of how the RbpjE89G allele affects sprouting angiogenesis in an experimentally tractable tissue like the postnatal retina.
Similar to the vasculature, patients with AOS can have a variety of cardiac pathologies, including atrial and ventricular septal defects, valve anomalies, aortic and pulmonic stenosis, coarctation of the aorta, and tetralogy of Fallot (2). Consistent with these findings, N1+/null Rbpj+/E89G,fl mice have abnormal cardiac morphology, most commonly membranous VSDs and dilated coronary vessels. The observed VSDs likely result from impaired growth or fusion of the endocardium with the cardiac neural crest–derived outflow tract septum (49). Dilated coronary vessels may be secondary to the heart failing (50) or due to aberrant patterning of vascular smooth muscle cells; the latter would be consistent with both mural cell patterning defects in patients with AOS (27) and the known role of Notch signaling in mural cell patterning (51–54). The lack of abnormal valve morphology in our mouse model is not surprising given that in mice, it is associated with modifiers such as diet (55, 56), which was not attempted in this study.
Although our study focused on defining the pathogenesis of cardiovascular defects, we were unable to similarly use our mouse model to assess the mechanisms underlying skin/scalp and limb defects, two widely regarded hallmarks of AOS in humans. In fact, throughout our mouse studies, we did not observe any obvious limb defects. However, scalp lesions were observed with one of the N1 alleles (N1gKO) that had considerable C57/BL6 in its genetic background, raising the possibility that this phenotype is sensitive to genetic background. Thus, comparative studies are needed using inbred mice carrying conditional N1 and RbpjE89G alleles to isolate the role of genetic background and test whether scalp lesions are due to defective N1 signaling in endothelial and/or other cell types.
Lastly, an unanswered question is how variants in RBPJ, which is the sole transcription factor downstream of all NOTCH receptors, cause an N1/DLL4 syndrome (AOS) but not an N2/JAG1 syndrome (Alagille syndrome) (25). Molecularly, RBPJ is thought to similarly interact with both NICD1 and NICD2, suggesting the RBPJ AOS variants should affect both N1- and N2-dependent processes. However, we found that the RbpjE89G allele in mice genetically interacts with N1 alleles to cause lethality and cardiovascular defects, whereas RbpjE89G and an N2-null allele were well tolerated in mice. Although additional studies are needed to assess whether RbpjE89G can affect some N2-sensitive cell types, these data suggest that the clinical importance of the RbpjE89G allele is due to its ability to preferentially compromise N1-dependent processes. Interestingly, comparative Notch signaling assays in cell culture revealed that ligand interactions with N2 generally produce more NICD molecules than N1 (19, 57). These studies suggest that the ratio of NICD coactivator to RBPJ transcription factor may contribute to the differential sensitivities of N1- versus N2-dependent processes to Rbpj AOS alleles. Importantly, the conditional mouse models generated in this study are ideally suited to assess how Rbpj AOS alleles affect N1- and N2-dependent processes during animal development.
-
Methods
Sex as a biological variable. AOS occurs in males and females without obvious bias (2, 5, 6). Nevertheless, we examined male and female mice and observed similar changes in viability in both sexes (see Supporting Data Values file for the sex of mice included in postnatal viability assays). Hence, we did not consider sex as a biological variable.
Structural modeling. The PyMOL Molecular Graphics System (version 3.0 Schrödinger, LLC) was used to visualize the structure of RBPJ bound to DNA (Protein Data Bank assembly 3BRG) (36). We used the PyMOL mutagenesis wizard to visualize the impact of AOS-associated mutations, selecting the rotamer for each variant that occurs most frequently in proteins. Discs represent pairwise overlap of atomic van der Waals radii. The color and size of each disc correlate with the amount of overlap. All human residue numbers correspond to the numbering used in isoform Q06330-1.
Protein purification. A pGEX-6P-1 plasmid encoding the conserved Rbpj core mouse residues 53–474 was used to generate each AOS variant through QuikChange mutagenesis using the primers in Supplemental Table 3. DNA constructs were confirmed by Sanger sequencing, and proteins were purified as previously described (34, 58). Protein concentrations were determined by measuring absorbance at 280 nm using a NanoDrop spectrophotometer. Protein purity was confirmed by SDS-PAGE with GelCode Blue staining (see Supplemental Figure 1B) per the manufacturer’s protocol (Thermo Fisher Scientific, 24590).
ITC. ITC experiments were performed as previously described (34). Briefly, purified RBPJ proteins were assessed for binding to the following: (a) an oligonucleotide sequence 5′–GGCACCGTGGGAAACTAGTG–3′ encoding a high-affinity RBPJ site (underlined); (b) a human NOTCH1 peptide consisting of residues 1754–1781; or (c) human SHARP residues 2776–2833. The NOTCH1 peptide was synthesized as previously described (34), and human SHARP residues were cloned into pSMT3 to produce protein with an N-terminal SMT3 and His tag as previously described (59). All proteins and DNA were dialyzed overnight in a buffer containing 50 mM sodium phosphate (pH 6.5) and 150 mM sodium chloride. Experiments were done in triplicate using a MicroCal VP-ITC. RBPJ plus DNA experiments were conducted at 10°C; RBPJ plus NICD/SHARP experiments were conducted at 25°C. Experiments were performed using 20 injections of 14 μL each. Heat-of-dilution experiments were conducted by injecting each ligand (DNA, NICD, or SHARP) in the syringe into a buffer-only solution in the cell. The heat-of-dilution experiment was subtracted from the experimental data before fitting. The raw data were analyzed using ORIGIN software and fit to a 1-site binding model. A 2-tailed t test was used to compare WT RBPJ with each variant; a P value less than 0.05 indicated a significant difference.
EMSAs. EMSAs were performed as described previously (16, 34, 60, 61). In brief, the 5′–CGAACGAGGCAAACCTAGGCTAGAGGCACCGTGGGAAACTAGTGCGGGCGTGGCT–3′ oligonucleotide containing an RBPJ site (underlined) was annealed to a complementary 5′IRDye-700 oligonucleotide 5′–AGCCACGCCCGCACT–3′. The duplex DNA was filled in using DNA polymerase I. Binding reactions were incubated for 20 minutes at room temperature, and protein-DNA complexes were separated by acrylamide gel electrophoresis. Gels were run for 2 hours at 150 V and imaged using a LI-COR Odyssey CLx scanner. Band intensity was quantified using Image Studio software (LI-COR Biotech LLC). Each experiment was performed in triplicate. A 1-way ANOVA with Tukey’s post hoc correction was used to compare WT RBPJ with each variant; a P value less than 0.05 indicated a significant difference.
Mice. Mice carrying RbpjS358R, RbpjE89G, and RbpjE89G,fl alleles were made in collaboration with the Cincinnati Children’s Hospital Medical Center Transgenic Animal and Genome Editing Facility (TAGE, RRID:SCR_022642) using CRISPR/Cas9 genome editing. For the RbpjS358R allele, we targeted cleavage to a site surrounding the S358 codon with the sgRNA 5′–TCCCTCATAGAACGTGTACTCGG–3′ and introduced a donor oligonucleotide 5′–ATCATTAGAACTGATAAAGCTGAGTATACG–3′ that substituted an arginine in place of S358 and introduced a DdeI restriction site for genotyping. For RbpjE89G and RbpjE89G,fl, we targeted cleavage to a site surrounding the E89 codon with the sgRNA 5′–AGTCTTACGGAAATGAAAAACGG–3′ and introduced a donor oligonucleotide 5′–CAGAAGTCATATGGGAATGGAAAA–3′ that substituted a glycine in place of E89 and introduced an NdeI restriction site for genotyping. RbpjE89G was made by editing WT CD1 mice; RbpjE89G,fl was made in outbred mice with existing flox sites surrounding exons 6 and 7 of the Rbpj gene (37). The genotypes of founder animals were confirmed using Sanger sequencing.
The other mouse lines used in this study included 3 N1 alleles: N1tm1Con (38) deletes genomic regions encoding amino acids 1056–2049, which includes the entire transmembrane region and Ankyrin repeats, and therefore is considered a constitutive null allele (N1null). The N1tm2Agt allele (39) was generated by incorporating loxP sites flanking the promoter and part of exon 1 followed by Cre recombination in the germline to make a constitutive N1-null allele referred to as N1gKO. The N1tm2Rko allele (44) was independently made in-house by inserting loxP sites in nearly identical sequences as Radtke et al. (39). We refer to this conditional allele as N1fl/fl. The other alleles used in this study were Rbpjnull (62), Rbpjfl/fl (37), N2LacZ (63), and Tie2-CreYwa (42). Offspring were genotyped using primers listed in Supplemental Table 2.
Timed embryonic harvest. Gestation was timed such that observation of a vaginal plug was considered E0.5. Pregnant dams were euthanized via CO2 inhalation followed by cervical dislocation, and the uterus was removed and placed into PBS on ice. Embryos were harvested and imaged with a Nikon SMZ 1500 stereoscope prior to collection of tissues. Specifically, the forelimbs, head, heart, and/or yolk sac were collected for analysis and placed into 4% paraformaldehyde (PFA) in PBS and incubated at 4°C overnight.
Western blotting. Single E10.5 Rbpj+/+ and RbpjE89G,fl/E89G,fl embryos were homogenized in 2x Laemmli sample buffer for Western blot analysis. Samples were sonicated and stored at –80°C. Protein extracts (whole embryos for RbpjE89G,fl/E89G,fl homozygotes, one-quarter embryos for WT controls) were run on a Bio-Rad 4%–20% Mini-PROTEAN TGX Stain-Free Precast Gel (catalog 456-8093) and transferred to a PVDF membrane via semidry transfer. The membrane was washed with water and then PBS before blocking with 0.5% casein in PBS for 1 hour at room temperature. The membrane was subsequently washed in PBS with 0.1% Tween-20, blocked in 0.5% casein with 0.05% Tween-20 in PBS (pH 7.4) for 1 hour at room temperature, and then incubated with antibodies against RBPJ (1:1,000, Cell Signaling Technology, 5313) and β-actin (1:2,000, LI-COR, 926-42212) overnight at 4°C. The membrane was washed in PBS with 0.1% Tween-20 and incubated with secondary antibodies (1:4,000 goat anti-rabbit IgG AF555, Invitrogen, A-21429; and 1:4,000 donkey anti-mouse IgG 680RD, LI-COR, 926-68072) at room temperature for 90 minutes. Finally, the membrane was washed in PBS with 0.1% Tween-20 and imaged using a Bio-Rad ChemiDoc imaging system. Band intensity was quantified using the Image Lab Software Suite (Bio-Rad), and RBPJ was normalized to β-actin levels.
Embryonic and postnatal heart assays. After overnight fixation in 4% PFA, E16.5 or postnatal hearts were washed 3 times for 5 minutes in PBS and submitted to the Integrated Pathology Research Facility for processing and embedding in paraffin (RRID:SCR_022637). Hearts were serially sectioned and either stained with H&E as described previously (64) or blocked and stained with 1:100 VE-cadherin (R&D Systems, AF1002). Stained heart sections were imaged using a Nikon NiE upright widefield microscope or Nikon A1R inverted confocal microscope.
Yolk sac vascular assays. E14.5 or E16.5 embryos were harvested and imaged within their yolk sacs from multiple angles with a Nikon SMZ 1500 stereoscope. Yolk sac vasculature was considered “reduced” if vitelline vessels were absent or markedly narrowed and/or if the visible capillary plexus extended over less than half of the yolk sac surface area. Yolk sac vasculature was scored by researchers blinded to genotype.
E10.5 embryos were fixed within their yolk sacs in 4% PFA in PBS for 30–60 minutes at room temperature. Embryos were washed 3 times for 5 minutes in PBS, dissected out of their yolk sacs, and reserved for genotyping. Empty yolk sacs were fixed in 4% PFA in PBS overnight at 4°C, washed 3 times for 5 minutes in PBT (PBS + 0.2% Triton X-100), blocked with 10% donkey serum in PBT for 2 hours at room temperature, and incubated with a rat anti-CD31 antibody (1:300, BD Biosciences, 553369) for 3 days at 4°C. Yolk sacs were washed 5 times for 15 minutes at room temperature with 2% normal donkey serum in PBT and incubated with a secondary antibody (1:300 donkey anti-rat AF647, Jackson ImmunoResearch Laboratories Inc., 712-605-153) for 2 days at 4°C. Yolk sacs were again washed 5 times for 15 minutes at room temperature and float-mounted in 1% agarose in coverslip-bottomed 48-well plates (Mattek, P48G-1.5-6). Tissue clearing was performed by adding 200 μL of EZClear (65) and incubating overnight prior to imaging with a Nikon A1R inverted confocal microscope. Image analysis and quantification were performed with AngioTool software (66). For calculating the percentage of vascular coverage, binaries were created for CD31-stained areas, and the relative coverage of the binaries compared with total image area was determined. For vascular diameter distributions, representative 400 μM × 400 μM areas were chosen and vessel diameters between all branch points were measured using the NIS-Elements measurements tool .
Embryonic skin vascular assays. Embryonic skin assays were performed essentially as previously described (67). In brief, PFA was removed from E14.5 forelimbs and heads by washing 3 times for 5 minutes in PBS. Tissues were transferred to 100% methanol (MeOH) for storage at –20°C. Using forceps, the skin was removed from the forelimbs and heads and rehydrated through a graded series of MeOH/PBT (PBS + 0.2% Triton X-100) washes. Skins were blocked with 10% donkey serum in PBT for 2 hours at room temperature and incubated with a rat anti-CD31 antibody (1:300, BD Biosciences, 553369) overnight at 4°C. Skins were then washed 5 times for 15 minutes at room temperature with 2% donkey serum in PBT and incubated with a secondary antibody (1:300 donkey anti-rat AF647, Jackson ImmunoResearch Laboratories Inc.,712-605-153) for 1 hour at room temperature. Skins were washed 5 times for 15 minutes at room temperature, mounted on slides, and imaged using a Nikon A1R inverted confocal microscope. Image analysis and quantification were performed with AngioTool (66) and Imaris software.
Statistics. Mouse viability was analyzed using the χ2 test for deviation from expected Mendelian ratios. Fisher’s exact test was used to determine whether the frequency of a phenotype differed between groups. Additional statistical tests are described in corresponding figure legends. For all statistical tests, a P value less than 0.05 indicated a significant difference.
Study approval. Animal experiments were carried out under protocols approved by the IACUC at Cincinnati Children’s Hospital Medical Center (protocols 2016-0105 and 2021-0086).
Data availability. All values underlying graphed data are available in the Supporting Data Values file.
-
Author contributions
RAK, RK, and BG conceptualized the study. AFS, KP, BC, HWL, CA, and EKG conducted formal analysis. BG acquired funding. AFS, KP, BC, RH, PG, ZY, BB, GM, HN, and EKG conducted the investigation. AFS, KP, BC, ZY, and EKG devised the methodology. RAK, RK, and BG were responsible for project administration. ZY, LS, and RAK provided resources. EKG, RAK, RK, and BG supervised the study. AFS and BG prepared the original draft. AFS, KP, BC, EKG, LS, RK, and BG reviewed and edited the manuscript.
-
Funding support
This work is the result of NIH funding, in whole or in part, and is subject to the NIH Public Access Policy. Through acceptance of this federal funding, the NIH has been given a right to make the work publicly available in PubMed Central.
- NIH (R01GM079428 to BG).
- National Science Foundation grant (2114950 to BG).
-
Acknowledgments
We thank Mei Xin for providing Tie2-CreYwa mice, Katherine Yutzey for expertise on heart morphology, and Elisa Boscolo and Yoh-suke Mukouyama for advice on vascular assays. This publication was made possible, in part, using the following Cincinnati Children’s Hospital Medical Center core facilities: Transgenic Animal and Genome Editing Facility [RRID:SCR_022642], Integrated Pathology Research Facility [RRID:SCR_022637], Genomics Sequencing Facility [RRID:SCR_022630], and Bio-Imaging and Analysis Facility [RRID: SCR_022628]. Parts of the figures and the graphical abstract were created with BioRender.com as indicated (https://BioRender.com/syfhnd9).
Address correspondence to: Brian Gebelein or Raphael Kopan, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, MLC7007, Cincinnati, Ohio, 45229. USA. Email: Brian.Gebelein@cchmc.org (BG); rafi.kopan@gmail.com (RK).
-
Footnotes
Conflict of interest: RAK is on the scientific advisory board of Cellestia Biotech AG and has received research funding from Cellestia Biotech AG for projects unrelated to this manuscript.
Copyright: © 2025, Solano et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2025;135(23):e187532.https://doi.org/10.1172/JCI187532.
-
References
- Adams FH, Oliver CP. Hereditary deformities in man: due to arrested development. J Hered. 1945;36(1):3–7.View this article via: CrossRef Google Scholar
- Meester JAN, et al. Elucidating the genetic architecture of Adams-Oliver syndrome in a large European cohort. Hum Mutat. 2018;39(9):1246–1261.
- Digilio MC, et al. Cardiovascular malformations in Adams-Oliver syndrome. Am J Med Genet A. 2015;167A(5):1175–1177.
- Southgate L, et al. Gain-of-function mutations of ARHGAP31, a Cdc42/Rac1 GTPase regulator, cause syndromic cutis aplasia and limb anomalies. Am J Hum Genet. 2011;88(5):574–585.
- Hassed SJ, et al. RBPJ mutations identified in two families affected by Adams-Oliver syndrome. Am J Hum Genet. 2012;91(2):391–395.
- Stittrich AB, et al. Mutations in NOTCH1 cause Adams-Oliver syndrome. Am J Hum Genet. 2014;95(3):275–284.
- Meester JA, et al. Heterozygous loss-of-function mutations in DLL4 cause Adams-Oliver Syndrome. Am J Hum Genet. 2015;97(3):475–482.
- Shaheen R, et al. Recessive mutations in DOCK6, encoding the guanidine nucleotide exchange factor DOCK6, lead to abnormal actin cytoskeleton organization and Adams-Oliver syndrome. Am J Hum Genet. 2011;89(2):328–333.
- Cohen I, et al. Autosomal recessive Adams-Oliver syndrome caused by homozygous mutation in EOGT, encoding an EGF domain-specific O-GlcNAc transferase. Eur J Hum Genet. 2014;22(3):374–378.
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233.
- Kovall RA, et al. The canonical Notch signaling pathway: structural and biochemical insights into shape, sugar, and force. Dev Cell. 2017;41(3):228–241.
- Falo-Sanjuan J, Bray SJ. Decoding the Notch signal. Dev Growth Differ. 2020;62(1):4–14.
- Morel V, Schweisguth F. Repression by suppressor of hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes Dev. 2000;14(3):377–388.
- Morel V, et al. Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr Biol. 2001;11(10):789–792.
- Chan SKK, et al. Role of co-repressor genomic landscapes in shaping the Notch response. PLoS Genet. 2017;13(11):e1007096.
- Kuang Y, et al. Enhancers with cooperative Notch binding sites are more resistant to regulation by the Hairless co-repressor. PLoS Genet. 2021;17(9):e1009039.
- Ilagan MX, et al. Real-time imaging of notch activation with a luciferase complementation-based reporter. Sci Signal. 2011;4(181):rs7.
- Gama-Norton L, et al. Notch signal strength controls cell fate in the haemogenic endothelium. Nat Commun. 2015;6:8510.
- Kuintzle R, et al. Diversity in Notch ligand-receptor signaling interactions. Elife. 2025;12:RP91422.
- MacGrogan D, et al. Notch and interacting signalling pathways in cardiac development, disease, and regeneration. Nat Rev Cardiol. 2018;15(11):685–704.
- Del Gaudio F, et al. Notch signalling in healthy and diseased vasculature. Open Biol. 2022;12(4):220004.
- Thambyrajah R, Bigas A. Notch signaling in HSC emergence: when, why and how. Cells. 2022;11(3):358.
- McLaren M, Butts J. Notch signaling in neurogenesis. Development. 2025;152(10):dev204589.
- Ramesh PS, Chu LF. Species-specific roles of the Notch ligands, receptors, and targets orchestrating the signaling landscape of the segmentation clock. Front Cell Dev Biol. 2023;11:1327227.
- Masek J, Andersson ER. The developmental biology of genetic Notch disorders. Development. 2017;144(10):1743–1763.
- Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 2017;97(4):1235–1294.
- Patel MS, et al. Abnormal pericyte recruitment as a cause for pulmonary hypertension in Adams-Oliver syndrome. Am J Med Genet A. 2004;129A(3):294–299.
- Swartz EN, et al. Vascular abnormalities in Adams-Oliver syndrome: cause or effect? Am J Med Genet. 1999;82(1):49–52.
- Pereira-Da-Silva L, et al. Clinical evidence of vascular abnormalities at birth in Adams-Oliver syndrome: report of two further cases. Am J Med Genet. 2000;94(1):75–76.
- Gale NW, et al. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc Natl Acad Sci U S A. 2004;101(45):15949–15954.
- Duarte A, et al. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 2004;18(20):2474–2478.
- De Zoysa P, et al. Murine model of cardiac defects observed in Adams-Oliver syndrome driven by Delta-Like Ligand-4 haploinsufficiency. Stem Cells Dev. 2021;30(12):611–621.
- Southgate L, et al. Haploinsufficiency of the NOTCH1 receptor as a cause of Adams-Oliver syndrome with variable cardiac anomalies. Circ Cardiovasc Genet. 2015;8(4):572–581.
- Gagliani EK, et al. A drosophila Su(H) model of Adams-Oliver Syndrome reveals cofactor titration as a mechanism underlying developmental defects. PLoS Genet. 2022;18(8):e1010335.
- Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79(2):273–282.
- Kovall RA, Hendrickson WA. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 2004;23(17):3441–3451.
- Han H, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14(6):637–645.
- Conlon RA, et al. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121(5):1533–1545.
- Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10(5):547–558.
- Meester JAN, et al. Overlapping but distinct roles for NOTCH receptors in human cardiovascular disease. Clin Genet. 2019;95(1):85–94.
- Krebs LT, et al. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 2004;18(20):2469–2473.
- Kisanuki YY, et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230(2):230–242.
- Srinivasan RS, et al. Lineage tracing demonstrates the venous origin of the mammalian lymphatic vasculature. Genes Dev. 2007;21(19):2422–2432.
- Yang X, et al. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev Biol. 2004;269(1):81–94.
- Huynh D, et al. Effective in vivo binding energy landscape illustrates kinetic stability of RBPJ-DNA binding. Nat Commun. 2025;16(1):1259.
- Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443.
- Noguera-Troise I, et al. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444(7122):1032–1037.
- Liu Z, et al. Notch1 loss of heterozygosity causes vascular tumors and lethal hemorrhage in mice. J Clin Invest. 2011;121(2):800–808.
- Neeb Z, et al. Cardiac outflow tract anomalies. Wiley Interdiscip Rev Dev Biol. 2013;2(4):499–530.
- Baba HA, Wohlschlaeger J. Morphological and molecular changes of the myocardium after left ventricular mechanical support. Curr Cardiol Rev. 2008;4(3):157–169.
- Feng X, et al. Patent ductus arteriosus in mice with smooth muscle-specific Jag1 deletion. Development. 2010;137(24):4191–4199.
- Chang L, et al. Differentiation of vascular smooth muscle cells from local precursors during embryonic and adult arteriogenesis requires Notch signaling. Proc Natl Acad Sci U S A. 2012;109(18):6993–6998.
- Manderfield LJ, et al. Notch activation of Jagged1 contributes to the assembly of the arterial wall. Circulation. 2012;125(2):314–323.
- Nadeem T, et al. Deficiency of Notch signaling in pericytes results in arteriovenous malformations. JCI Insight. 2020;5(21):e125940.
- Garg V, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437(7056):270–274.
- Joll JE, et al. Genetic ablation of serotonin receptor 2B improves aortic valve hemodynamics of Notch1 heterozygous mice in a high-cholesterol diet model. PLoS One. 2020;15(11):e0238407.
- Liu Z, et al. The extracellular domain of Notch2 increases its cell-surface abundance and ligand responsiveness during kidney development. Dev Cell. 2013;25(6):585–598.
- Friedmann DR, et al. RAM-induced allostery facilitates assembly of a notch pathway active transcription complex. J Biol Chem. 2008;283(21):14781–14791.
- VanderWielen BD, et al. Transcriptional repression in the Notch pathway: thermodynamic characterization of CSL-MINT (Msx2-interacting nuclear target protein) complexes. J Biol Chem. 2011;286(17):14892–14902.
- Uhl JD, et al. Comparing anterior and posterior Hox complex formation reveals guidelines for predicting cis-regulatory elements. Dev Biol. 2010;343(1-2):154–166.
- Kuang Y, et al. Enhancer architecture sensitizes cell specific responses to Notch gene dose via a bind and discard mechanism. Elife. 2020;9:e53659.
- Oka C, et al. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 1995;121(10):3291–3301.
- Skarnes WC, et al. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc Natl Acad Sci U S A. 1995;92(14):6592–6596.
- Cardiff RD, et al. Manual hematoxylin and eosin staining of mouse tissue sections. Cold Spring Harb Protoc. 2014;2014(6):655–658.
- Hsu CW, et al. EZ Clear for simple, rapid, and robust mouse whole organ clearing. Elife. 2022;11:e77419.
- Zudaire E, et al. A computational tool for quantitative analysis of vascular networks. PLoS One. 2011;6(11):e27385.
- Li W, Mukouyama YS. Whole-mount immunohistochemical analysis for embryonic limb skin vasculature: a model system to study vascular branching morphogenesis in embryo. J Vis Exp. 2011;51:2620.
- Adams FH, Oliver CP. Hereditary deformities in man: due to arrested development. J Hered. 1945;36(1):3–7.
-
Version history
- Version 1 (October 7, 2025): In-Press Preview
- Version 2 (December 1, 2025): Electronic publication



Copyright © 2026 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)












