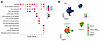Advertisement
Research ArticleImmunologyOncology
Open Access |  10.1172/JCI190778
10.1172/JCI190778
Tissue-specific antitumor NK cell subsets identified in colorectal cancer liver metastases express candidate therapeutic targets
Joanna Mikulak,1 Domenico Supino,2 Paolo Marzano,1,3 Sara Terzoli,1,4 Roberta Carriero,5 Valentina Cazzetta,1 Rocco Piazza,6 Elena Bruni,3 Paolo Kunderfranco,5 Alessia Donato,2 Sarah Natalia Mapelli,2 Roberto Garuti,2,4 Silvia Carnevale,7 Francesco Scavello,2 Elena Magrini,2 Jelena Zeleznjak,2,8 Clelia Peano,9,10 Matteo Donadon,11 Guido Costa,12 Guido Torzilli,4,12 Alberto Mantovani,2,4,13 Cecilia Garlanda,2,4 and Domenico Mavilio1,3
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Mikulak, J. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Supino, D. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by
Marzano, P.
in:
PubMed
|
Google Scholar
|

1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Terzoli, S. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Carriero, R. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Cazzetta, V. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by
Piazza, R.
in:
PubMed
|
Google Scholar
|

1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by
Bruni, E.
in:
PubMed
|
Google Scholar
|

1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by
Kunderfranco, P.
in:
PubMed
|
Google Scholar
|

1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Donato, A. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Mapelli, S. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Garuti, R. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Carnevale, S. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Scavello, F. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Magrini, E. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Zeleznjak, J. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Peano, C. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Donadon, M. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Costa, G. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by
Torzilli, G.
in:
PubMed
|
Google Scholar
|

1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Mantovani, A. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by Garlanda, C. in: PubMed | Google Scholar
1Unit of Clinical and Experimental Immunology and
2Unit of Experimental Immunopathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
3Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
4Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
5Bioinformatics Unit, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
6Department of Medicine and Surgery, University of Milan-Bicocca, Monza, Italy.
7Unit of Innate Immunity in Inflammation and Cancer, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
8Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia.
9Institute of Genetics and Biomedical Research, National Research Council, Rozzano, Milan, Italy.
10Human Technopole, Milan, Italy.
11Department of Health Sciences, Università del Piemonte Orientale, Novara, Italy; Department of General Surgery, University Maggiore Hospital Della Carità, Novara, Italy.
12Unit of Hepatobiliary and General Surgery, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy.
13William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Find articles by
Mavilio, D.
in:
PubMed
|
Google Scholar
|

Authorship note: JM and DS contributed equally to this work and have been designated as co–first authors. DM and CG contributed equally to this work and have been designated as co–last authors.
Published October 28, 2025 - More info
J Clin Invest. 2025;135(24):e190778. https://doi.org/10.1172/JCI190778.
© 2025 Mikulak et al. This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Received: December 30, 2024; Accepted: October 21, 2025
-
Abstract
Liver metastases are relatively resistant to checkpoint blockade immunotherapy. The hepatic tissue has distinctive features including high numbers of NK cells. It was therefore important to conduct in-depth single-cell analysis of NK cells in colorectal cancer liver metastases (CRLMs) with an effort to dissect their diversity and to identify candidate therapeutic targets. By combining unbiased single-cell transcriptomic with multiparametric flow cytometry analysis, we identified an abundant family of intrahepatic CD56bright NK cells in CRLMs endowed with antitumor functions resulting from specific transcriptional liver programs. Intrahepatic CD56bright and CD56dim NK lymphocytes expressed unique transcription factors (IRF8, TOX2), a high level of chemokines, and targetable immune checkpoints, including CXCR4 and the IL-1 receptor family member IL-1R8. CXCR4 pharmacological blocking and an anti–IL-1R8 mAb enhanced the effector function of CRLM NK cells. Targeting the diversity of liver NK cells and their distinct immune checkpoint repertoires is key to optimize the current immune therapy protocols in CRLM.
-
Introduction
Colorectal cancer (CRC) is the third most frequent human malignancy and shows high rates of hepatic metastasis. Indeed, approximately 50% of CRC patients develop liver metastases (CRLMs) either as synchronous or metachronous disease (1, 2). Although the clinical outcome of CRLM remarkably improved over the past 20 years, we still lack a curative approach. The administration of neoadjuvant chemotherapies (na-CHTs) combined with biologic treatment followed by surgical resection represents the main available treatment, with a 5-year overall survival between 20% and 45% and a recurrence of 60% (3–5).
Several immune checkpoint (IC) inhibitors have been approved for clinical use and achieved encouraging results in a variety of solid tumors (6). However, CRLM patients showed poor clinical benefits from immunotherapies that included anti–PD-1/PD-L1 and/or anti-CTLA4 (5, 7–10), probably owing to the unique liver tumor microenvironment (11, 12). In addition, immunotherapeutic approaches are associated with favorable prognosis mainly in microsatellite instability–high/mismatch repair–deficient CRC tumors displaying high overall mutation burden, which represent about 3% of CRLMs (13–15). This is not the case for microsatellite-stable/mismatch repair–proficient CRC tumors (16–18). Therefore, exploring the heterogeneity of tumor-infiltrating immune cells and identifying cell type–specific ICs is key to identify the best responses to immunotherapies and to improve the clinical outcomes of CRLM patients.
The liver is considered a preferential tissue for natural killer (NK) cell residency. In humans, NK cells can reach up to 50% of intrahepatic lymphocytes and show a broad spectrum of functions by regulating the dynamic balance between immune tolerance and immune surveillance (19–22). Since their discovery in the mid-1970s, NK cells have been valued for rapid recognition and clearance of cancer cells in the absence of antigen specificity (23). Human NK cells are divided into 2 main subsets defined on the basis of their differential expression of CD56 and CD16, namely CD56brightCD16– (CD56bright) and CD56dimCD16+ (CD56dim) NK cells (24, 25). Intrahepatic CD56bright cells represent the major NK cell subset characterized by constitutive expression of the CXCR6 receptor and the tissue residency marker CD69 (26, 27). NK cell cytotoxicity and secretion of interferon-γ (IFN-γ) are controlled by a dynamic balance exerted by an array of inhibitory receptors (iNKRs) and activating receptors (aNKRs) differentially expressed on the cell surface of NK cells (28).
Several studies reported the presence at high frequencies of tumor-infiltrating NK cells within primary and metastatic liver malignancies, including hepatocellular carcinoma and CRLMs (22, 29–32). However, the relevance and clinical impact of tissue-resident NK cell immune surveillance against CRLMs are still largely unknown and being debated (21).
The present study had the aim of dissecting the functional heterogeneity of matched circulating and intrahepatic NK cells isolated from CRLM patients undergoing surgical resection of metastatic lesions, as well as investigating the expression of ICs and other potential targets of immunotherapy. By combining single-cell transcriptional analysis of NK and CD45+ cells with multiparametric flow cytometry and functional assays, we show here that intrahepatic CD56bright NK cells in CRLMs are mature and potent tissue-resident antitumor effectors expressing a unique transcriptional profile that makes them distinguishable from their circulating counterparts and from both peripheral and intrahepatic CD56dim NK cells. Moreover, the liver metastatic niche is characterized by the expression of ICs on several NK cell subsets. Among these, we focused on the IL-1 receptor family member IL-1R8, which we previously identified as an IC that negatively regulates IL-18–dependent NK cell activation, involved in resistance to hepatic carcinogenesis and hematogenous CRLM (31). We found that IL-1R8 was ubiquitously expressed by intrahepatic NK cells, and its targeting with a blocking monoclonal antibody (mAb) improved NK cell antitumor effector functions. In addition, CXCR4 emerged as a potential therapeutic target, as suggested by the increased NK cell production of IFN-γ after its blocking with the antagonist plerixafor.
-
Results
Dissection of intrahepatic NK cell heterogeneity in CRLM by single-cell RNA sequencing. To decipher the heterogeneity of intrahepatic NK cells, single-cell RNA sequencing (scRNA-Seq) was performed. FACS-sorted NK cells (Lin–CD56+, excluding CD3+, B, and myeloid cells) were isolated from CRLM and peripheral blood (PB) samples from 3 patients (2 male and 1 female) undergoing limited surgical liver resection for synchronous CRLM disease after na-CHT treatment (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI190778DS1). Samples were collected from the peritumor (PT) and invasive margin (IM) areas of CRLM. The integrated transcriptomic profiles of 14,091 PT and 9,208 IM liver NK (NKLR) cells were interrogated by nonlinear dimension reduction with uniform manifold approximation and projection (UMAP) (Figure 1A and Supplemental Figure 1C). In total, 18 liver cell clusters (cLR) were obtained. PT and IM NKLR cells were equally distributed among clusters with minor differences between individual patients (Figure 1B and Supplemental Figure 1D) and were thus merged for subsequent analyses. Contaminating clusters enriched in CD3+ lymphocytes and myeloid and B cells (c10LR, c12–17LR) represented less than 0.07% of total events and were excluded from further analysis (Figure 1C and Supplemental Figure 1E).
 Figure 1
Figure 1Single-cell RNA-Seq analysis of liver NK cells from invasive margin and peritumor CRLM tissue sites. (A) UMAP projection of liver NK (NKLR) cells from invasive margin (IM; n = 9,208) and peritumor (PT; n = 14,091) of 3 CRLM patients. CD56bright clusters are circled in green, CD56dim in red. (B) Bar plot showing relative frequency of clusters in IM and PT (normalized per tissue). (C) Left: Dot plot of NCAM1, FCGR3A, and CD3G expression. Right: Kernel density of NCAM1 and FCGR3A on UMAP plot. (D) Violin plots of selected genes across NKLR cell clusters, ordered according to CD56bright or CD56dim.
Unsupervised analysis subdivided NKLR cells into two CD56dim and CD56bright macro-clusters that differed in the expression of CD16 (FCGR3A) and CD56 (NCAM1) (Figure 1C, Supplemental Figure 1F, and Supplemental Table 2) (33, 34). CD56dim and CD56bright NKLR cells included 3 clusters (c3–4LR, c6LR) and 8 clusters (c0–2LR, c5LR, c7–9LR, c11LR), respectively. As expected, the majority of the NKLR fraction (69%) was represented by CD56bright cells (Figure 1, A and B) (26). Differentially expressed gene (DEG) analysis confirmed the liver-residency profile of CD56bright NKLR cells, characterized by coexpression of CXCR6, EOMES, and CD69 and lack of CD52 (Figure 1D and Supplemental Figure 2) (21). In addition, CD56bright NKLR cells shared high expression of the chemokines XCL1 and XCL2, the cytokine receptor IL2RB, the costimulatory molecule ICAM1, and a peculiar coexpression of transcription factors (TFs) both inhibiting (TCF7) and promoting (IRF8, TOX2) NK cell maturation (35–39). On the other hand, CD56dim NKLR cells expressed FGFBP2, the memory NK cell marker CD52, and TFs such as PRDM1, KLF2, and the CX3CR1 receptor, which is involved in NK cell migration from PB to peripheral tissues (40–42), thus suggesting they are not a resident population (Figure 1D). Although we cannot entirely exclude the presence of intravascular NK cells, given the highly perfused nature of the liver, the transcriptional profile of CD56dim NKLR cells strongly suggests peripheral recruitment or transient retention.
Pseudotime analysis was applied to unravel the developmental trajectories between NKLR cell clusters in CRLM. The pseudotime results by Monocle2 showed CD56bright and CD56dim cells along 3 separated branches (Figure 2, A and B, and Supplemental Figure 3A). 97.2% of CD56dim NK cells were located on the right branch; in contrast, the majority of CD56bright NK cells were distributed among the left (62.7%) and upper (28.1%) branches (Figure 2B). Importantly, we observed a minimal developmental relationship between CD56bright and CD56dim NK cells (9.1%) (Figure 2B), suggesting that a direct transition between the 2 subsets may be less evident in this setting. To validate this observation, pseudotime trajectories were also inferred using the Slingshot algorithm (Supplemental Figure 3B). This analysis confirmed the relative independence of intrahepatic CD56bright and CD56dim NK cells and highlighted the enrichment of CD56bright clusters with distinct functional profiles within the CRLM immune environment.
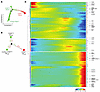 Figure 2
Figure 2Pseudotime analysis of CD56bright and CD56dim NKLR cells. (A) Monocle2 pseudotime trajectory of NKLR cells. CD56bright and CD56dim cells are shown in green and red, respectively. (B) Relative frequency of CD56bright (green) and CD56dim (red) cells across branches, normalized within each branch. (C) Heatmap of selected driver genes along pseudotime. Cells are ordered left to right; colors indicate scaled (z score) expression.
Highly proliferative cells in CD56bright c8LR and c7LR expressing hallmark genes of DNA replication (e.g., TUBB, MIKI67, STMN1) were located at the starting root node of the CD56bright cell branch (Figure 2, A–C, and Supplemental Figure 3, A and C). These were associated with a liver tissue-residency profile, as suggested by the expression of EOMES and CXCR6, and terminal maturation markers such as TOX2 and CD244 (Figure 1D, Figure 2C, and Supplemental Figure 3C), indicating their high transcriptional dynamics. From these, we observed a progression toward a full effector stage following the CD56bright cell fate of c0–2LR, as suggested by pseudotime-dependent DEGs including cytokines (e.g., XCL1, IL16, and TNF), effector function molecules (e.g., GZMB, GZMK, and PRF1), and TFs (e.g., EOMES and TOX2) (Figure 2C and Supplemental Figure 3C). Finally, we further identified CD56bright c9LR and c11LR clusters, characterized by more advanced pseudotime positions, showing high expression of adaptive-like (KLRC2, CD3E) (43) and mitochondrial-active (e.g., MTRNR2L1/8/12/18) signature genes, respectively (Figure 1D, Figure 2C, Supplemental Figure 2, and Supplemental Figure 3, A and C).
On the other hand, CD56dim NK cells showed lower transcriptional heterogeneity as highlighted by the expression of terminally mature NK cell markers (KIRs, B3GAT1, CD52, TBX21, KLRG1, FGFBP2, NKG7, CD226), cytotoxic mediators (GZMB, GZMH, PRF1, GNLY, CTSW), and the tissue-homing receptor CX3CR1 (Figure 1D, Figure 2C, and Supplemental Figure 2).
Overall, pseudotime analysis revealed that intratumoral hepatic CD56bright and CD56dim NK cells followed largely independent trajectories with minimal overlap, with CD56bright cells displaying multiple intermediate states, whereas CD56dim cells predominantly occupied terminally differentiated states with limited heterogeneity.
Functional status and immune checkpoint profile of liver NK cells in CRLM. We then performed Gene Ontology (GO) enrichment analysis of DEGs for biological processes to elucidate pathways associated with intrahepatic NK cell clusters. GO enrichment analysis highlighted a strong association of NKLR cells with “cell killing” (Figure 3A). “NK cell–mediated cytotoxicity” mainly correlated with the CD56dim transcriptomic profile. Cell cycle and proliferation pathways were associated with c7LR and c8LR CD56bright clusters, whereas “cellular oxidant detoxification” and electron transport chain pathways were enriched in c11LR, as marked by mitochondrial genes. “Type I IFN production” was exclusively observed in CD56dim cells, whereas pathways of IFN-γ production were shared by CD56bright and CD56dim NKLR cells. In particular, we identified 2 clusters, CD56bright c5LR and CD56dim c6LR, associated with a distinctive IFNGhigh transcriptomic profile (Figure 3B), suggesting their effector activation. IFNGhigh cell subsets showed higher expression of heat shock proteins (HSPs) (e.g., HSPA1A, HSPA1B, HSPB1), chaperone and co-chaperone for HSP (DNAJA1, CHORDC1), and signaling molecules such as JUN, GIMAP4, and DUSP1 (Figure 4, A and B). Similar results were observed both in PT and IM compartments (Supplemental Figure 4A).
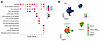 Figure 3
Figure 3Functional enrichment and IFNG expression in NKLR cells. (A) Dot plot of GO biological process terms (q ≤ 0.01) enriched in intrahepatic NKLR cell clusters [DEGs: FDR ≤ 0.05, log(fold change) > 0.25]. Dots are colored by EnrichFold and sized by adjusted P value. (B) Kernel density of IFNG expression on the UMAP plot (top). The clustering of cells is shown at the bottom, with clusters grouped according to their transcriptional features.
 Figure 4
Figure 4Comparison of liver CD56bright and CD56dim NKLR cells. (A) Volcano plot of DEGs in CD56bright c5LR versus c0–1–2LR (left) and CD56dim c6LR versus c3–4LR (right). (B) Heatmap of IFNGhigh signature gene expression across NKLR clusters. (C) Violin plots of selected immune checkpoint genes.
To decipher the mechanisms that regulate these IFNG-expressing cells conserved across CD56bright and CD56dim NKLR, we investigated the expression of ICs. CD56bright and CD56dim cell subsets were distinguished for their IC pattern (Figure 4C). KLRC1, TIGIT, CD96, and CD160 were mainly associated with the CD56bright phenotype. In addition, ENTDP1 (CD39) was expressed by cycling CD56bright cells in c7–8LR. LAG3 identified adaptive-like (c9LR) CD56bright cells. In contrast, CD56dim cells were marked by HAVCR2 (TIM-3) and KLRG1, and higher expression of specific KIRs (Supplemental Figure 4B). We found low expression of PDCD1 (PD-1) and CTLA4 in all NKLR cells. Notably, SIGIRR (IL-1R8), which we previously identified as an NK cell IC (31), was ubiquitously expressed by all NKLR cell clusters. In addition, lower expression of CXCR4, an unconventional IC of liver NK cells (44), was observed in the IFNGhigh c5LR and c6LR (Figure 4C), as also shown in the heatmap (Figure 4B).
To precisely define the contribution of CRLM microenvironment to the orientation of intrahepatic NK cells, we harmonized scRNA-Seq data from healthy liver samples (45) (see Methods). In the healthy liver, we identified CD56bright, CD56dim, and cycling NK cell subsets, with a clear separation between the CD56bright and CD56dim populations (Figure 5, A and B, and Supplemental Table 2) (26, 33, 34). This distinction mirrors what we observed in CRLM samples, as illustrated by UMAP embedding annotated with phenotype-specific gene signature scores (Figure 5B).
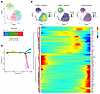 Figure 5
Figure 5Healthy hepatic NK cells. (A) Top: UMAP projection of integrated NK cells (n = 5,760) from 5 healthy donors. Bottom: The bar plot shows the cluster frequencies normalized on total cells. (B) Kernel density of CD56bright, CD56dim, and cycling signature scores on UMAP. (C) Left: Pseudotime trajectory of hepatic NK cells (colored by cluster). Right: Heatmap of selected driver genes ordered by pseudotime, with scaled (z score) expression shown by color.
As in the pathological condition, pseudotime analysis in healthy liver also revealed only a minimal developmental connection between CD56bright and CD56dim NK cells (Figure 5C), with cycling NK cells located at the starting root of the trajectory. However, when we projected the pathological pseudotime ordering genes (Figure 2C) onto the healthy dataset (Figure 5C), many of them were absent (e.g., GZMA, TNF, CD244 [2B4], IL16, PRF1), indicating that the pathological milieu shapes the developmental trajectory of NK cells.
To investigate the NK cell subset–specific transcriptional changes induced by CRLM, we performed DEG analysis (Figure 6, A and B). Volcano plots highlighted that both CD56bright and CD56dim NK cells exhibited features of activation, including the upregulation of transcriptional programs mediated by AP-1 and NF-κB, as well as activation of MAPK signaling pathways. In both subsets, we observed increased expression of IFNG, cytotoxic effectors (GZMB, GNLY, PRF1), activating receptors (FCGR3A, NKG7), and transcriptional regulators such as TOP2B, BRD4, and POLR2A. Notably, CD56bright NK cells showed elevated expression of effector-associated genes such as EOMES, FAS, TOX, CRTAM, and TNFRSF9. Similarly, CD56dim NK cells upregulated the effector-associated gene CD226 (DNAM-1) and differentiation markers such as B3GAT1 (CD57) along with KIRs and TGFB1, TGFBR2, and TGFBR3, important regulators of NK cell activity (46). We also observed changes in the expression of immune checkpoint molecules: CD56bright NK cells expressed higher levels of TIGIT and ENTPD1, while CD56dim NK cells upregulated KLRC1 and LAIR2, supporting our findings of subset-specific immune regulation (Figure 4C).
 Figure 6
Figure 6Comparison of CD56bright and CD56dim NK cells in CRLM versus healthy liver. Volcano plots of DEGs for CD56bright clusters (A) and CD56dim clusters (B) in CRLM versus healthy liver.
Together, these data demonstrate that CRLM profoundly reshapes the transcriptional landscape of both CD56bright and CD56dim NK cells, fostering an effector-like phenotype marked by a strong IFNG signature alongside finely tuned immune checkpoint modulation. While both subsets exhibit activation features, they preserve their distinct transcriptional and phenotypic identities, challenging the notion that effector functions in intrahepatic NK cells are confined to the CD56dim population.
Ligand-receptor interactions shaping NKLR crosstalk in CRLM. The identification of an IFNG-associated signature within intrahepatic CD56bright and CD56dim cells suggests that the activation of this immune pattern might be elicited by common signals in CRLM patients. We thus investigated how intrahepatic CD56bright and CD56dim NK cell subsets interact with different components of the CRLM immune microenvironment. We used NicheNet, an algorithm that infers ligand-target activities on the basis of their expressed genes (47). First, scRNA-Seq analysis of intrahepatic CD45+ cells of the same patients was used to annotate leukocyte clusters (Figure 7A) (48). We set the three NK cell clusters emerging from CD45+ cell analysis (CD56bright, CD56dim, and cycling NKLR cells) as “receiver” populations, focusing our analysis on the top-scored predicted NK ligands expressed in the “sender” myeloid and lymphoid cells (Figure 7B). Intrahepatic CD56bright and CD56dim NK cells shared most of their ligands upon interaction with either lymphoid or myeloid compartments. In contrast, cycling intrahepatic NK cells established specific ligand-target interactions. Ligands interacting with intrahepatic CD56bright and CD56dim NK cells, including IFNG, TGFB1, CD40LG, and the cytokines IL2, IL21, and IL23A, were largely expressed in lymphoid cells. IL18, IL1B, IL27, PTGS2, and IL15 were mainly expressed by myeloid senders (Figure 7C). Predicted ligands were involved in intrahepatic NK cell–mediated killing (e.g., PRF1, GZMs, FASLG), cytokine secretion (e.g., CCL3–5, XCL1, IRFs), activation and maturation (e.g., JUN, NFKBIA, MAFF, TBX21, NCR3), and cell migration (e.g., ICAM1–2, ITGAM). These results suggested that intrahepatic NK cell cytotoxicity was regulated by IL2 and TGFB1 expressed by CD4+ T cells, type 3 innate lymphoid cells (ILC3s), and γδ T cells (Figure 8A). In addition, we found IL18 as a key IFNG-upstream ligand for both intrahepatic CD56bright and CD56dim NK cells in CRLM patients. Kupffer cells (KCs) and type 2 conventional dendritic cells (cDC2s) were identified as a major source of IL18. Focusing on IFNG-expressing cells, we found that intrahepatic NK cell ligands were broadly expressed among lymphocytes and myeloid cells (Figure 8, B and C). In contrast, CXCL12, the ligand for CXCR4 receptor, was uniquely expressed by KCs, suggesting they regulate the CXCL12/CXCR4 axis in the CRLM milieu.
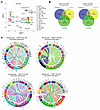 Figure 7
Figure 7Intercellular communication in the immunological milieu of CRLM. (A) UMAP projection of intrahepatic CD45+ cells colored by cell family. (B) Venn diagrams of common and specific ligands from lymphoid (left) and myeloid (right) cells interacting with CD56bright, CD56dim, and cycling NK cells. (C) Chord plots of the top 100 predicted interactions (NicheNet) for CD56bright (top) and CD56dim (bottom) NK cells, ranked by regulatory potential. In each circular plot, one arc contains the ligands (derived from lymphoid [left] or myeloid [right] cells) and the opposite arc contains the downstream NK target genes. Directional chords connect each ligand to its predicted NK target genes, with the pointed end of each chord indicating the direction of regulation. Bars above the ligand arc represent the proportion of downstream target genes regulated by each ligand.
 Figure 8
Figure 8Ligand expression and IFNG interactions in intrahepatic CD45+ cells. (A) Dot plot of selected ligands in intrahepatic CD45+ populations, stratified by biological function. (B) Dot plot of ligands from intrahepatic CD45+ populations interacting with IFNG+ NK cells, stratified by function. (C) Feature plot of IFNG expression on intrahepatic CD45+ cells.
Checkpoint landscape defines NK cell identity in CRLM. Using the scRNA-Seq dataset of CD45+ cells in CRLMs (Supplemental Figure 5), we conducted a parallel evaluation of NK cells and other lymphocyte types. Peculiarly, most lymphocyte clusters showed high expression of tissue-affinity genes such as EOMES, CXCR6, and CD69, especially mucosal-associated invariant T (MAIT), CD8+, and γδ T cells. In addition, CD3+ T cells displayed effector function molecules described in intrahepatic CD56bright NK cells, but not in CD56dim clusters. For instance, γδ T cells and ILC3s are characterized by high levels of XCL1 and XCL2. Furthermore, GZMK has been detected in several clusters but not in CD56dim NK cells. Notably, MAIT, CD8+, and γδ T cells markedly contribute to TNF and IFN-γ production, while CD56dim NK cells are the predominant cytotoxic population, as suggested by the combination of GZMB, GNLY, and PRF1 levels. The evaluation of TFs has further revealed that TOX2, a strong terminal maturation/exhaustion marker in lymphocytes, is unexpectedly characteristic of CD56bright NK cells and ILC3s.
IC analysis revealed a shared expression of regulatory molecules such as CD96, SIGIRR, and CXCR4, across distinct immune lymphocyte types, suggesting their common involvement in orchestrating immune regulation within CRLM. However, combinatorial IC pattern analysis uncovered cell type–specific IC signatures. For instance, NKLR cells differed markedly from T cells by lacking expression of LAG3, PDCD1, and KLRG1. In contrast, NKLR cell subsets displayed elevated expression of KLRC1, ENTPD1, HAVCR2, and CD160, with expression levels varying between CD56dim and CD56bright NKLR cells. These data point to a finely tuned, subset-specific regulatory landscape that defines NK cell identity and function within the CRLM microenvironment. These distinct checkpoint architectures may pave the way for subset-tailored immunotherapeutic strategies aimed at reinvigorating NK cell–mediated antitumor activity in CRLM.
Dissection of PB NK cell heterogeneity in CRLM. In parallel, we performed scRNA-Seq analysis of NK cells isolated from matched PB samples. Based on transcriptomic profiles, a total of 12,820 PB NK (NKPB) cells were projected by UMAP. The unsupervised analysis identified 14 cell clusters (c0–13PB) with a similar relative frequency among each donor (Figure 9A and Supplemental Figure 6A). The lack of FCGR3A and NCAM1 expression together with a myeloid cell signature prompted us to exclude c9PB from further analysis (Figure 9B and Supplemental Figure 6B). Two main CD56bright and CD56dim phenotypes were annotated by hierarchical clustering and by transcriptional profiles (Figure 9, C and D, Supplemental Figure 6, C and D, and Supplemental Table 2) (33, 34). CD56bright NKPB cells included 3 clusters (c8PB, c4PB, c13PB) that represented a minority (9.6%) of total NKPB cells. CD56dim NKPB cells were grouped into 4 major (c0–3PB) and 5 minor (c5–7PB, c11–12PB) clusters. In contrast to their intrahepatic counterparts, CD56bright and CD56dim NKPB cells showed interrelated differentiation states with continuous progression. The pseudotime trajectory began with the CD56bright c8PB cluster expressing immature NK cell markers such as COTL1, SELL, CD44, CD2, XCL1, XCL2, and GZMK (Figure 9, E and F, and Supplemental Figure 7, A–C). Transitional states between CD56bright and CD56dim phenotypes were observed in clusters c4PB and c2PB, as suggested by increased effector potential (e.g., GZMs, FCGR3A, CTSW, KLRB1, NKG7, TBX21, NCR3) and lower expression of CD56bright markers (e.g., XCL1, XCL2, CD44, GZMK, NCR1) in comparison with c8PB (Figure 9, D and F, and Figure 10A). Likewise, NK cells in c2PB showed a less mature phenotype in comparison with the other CD56dim clusters (Figure 10B). We further detected increased expression of CX3CR1 in c1PB indicating preferential migration of mature CD56dim cells to the CRLM microenvironment (Figure 1D).
 Figure 9
Figure 9Single-cell RNA-Seq analysis of PB NK cells in CRLM patients. (A) UMAP projection of integrated PB NK (NKPB) cells (n = 12,820) from 3 patients, showing CD56bright (green) and CD56dim (red) clusters. (B) Dot plot of NCAM1, FCGR3A, and CD3G expression. (C) Overlaying clustering tree of PC3 and PC4 showing CD56bright and CD56dim cells at resolutions 0.2 (pink) and 0.7 (blue). (D) Violin plots of selected genes characteristic of CD56bright (green) and CD56dim (red). (E) Pseudotime trajectory of NKPB cells colored by cell family (top) or pseudotime value (bottom). (F) Heatmap of selected genes along pseudotime.
 Figure 10
Figure 10Differential gene expression in NKPB cell clusters. (A) Volcano plot of DEGs for CD56bright c8PB versus c4PB. (B) Heatmap of selected DEGs from CD56bright c8PB versus c13PB, and CD56dim c1–7PB and c10–12PB clusters. Expression values are zero-centered and scaled per gene.
CD56bright (c13PB) and CD56dim (c5–7PB and c10–12PB) cells showed tumor-related transcriptomic profiles (Figure 10B). In particular, c13PB showed tissue residency and effector genes previously identified in intrahepatic CD56bright cells (e.g., CXCR6, FASLG, IRF8, TIGIT, CD160, TOX2, CHORDC1), thus suggesting their egression from the liver to the blood. CD56dim cells in c6PB expressed a family of mitochondrial genes (e.g., MTRNR2L8/12) (49, 50). CD56dim NKPB cells in c5PB and c11PB were characterized by an apoptotic (e.g., BAX, BBC3, MDM2) and a proliferating (e.g., MIK67, PCNA, STMN1) phenotype, respectively. CD56dim c12PB cells, similarly to c13PB, mirrored a liver signature (i.e., IFNG, DNAJB1, HSPA1A, HSPB1). Finally, CD56dim c10PB showed an activated phenotype (e.g., IRF8, IER5, IER2, NFKBIA, RELB) and c7PB an adaptive-like profile as indicated by higher expression of CD3E, CD3G, CD52, and LAG3 and lower expression of FCER1G (FcεRγ), ZBTB16, SYK, SH2D1B (EAT2), and KLRB1 (43, 51).
Multiparametric flow cytometry profiling of NK cells in CRLM patients. scRNA-Seq analysis highlighted a specific IC pattern in intrahepatic NK cells depending on tissue residency, effector stage, response to CRLM, and likely egress of intrahepatic NK cells to bloodstream. We next proceeded to validate the NK cell expression of ICs by multiparametric flow cytometry of liver samples and PB isolated from CRLM patients and compared with PB of healthy donors (HDs). We confirmed that intrahepatic NK cells expressed high levels of TIGIT, CD39, NKG2A, and CD96, although with high heterogeneity among patients. Notably, TIGIT and CD39 were mainly enriched on CD56bright NKLR cells from CRLM patients (Figure 11A), and in line with the transcriptional profiles, the expression of these two ICs was higher on CD56bright NKLR cells compared with their PB counterparts. In contrast, CD56bright NKPB cells presented higher expression of NKG2A and CD96 compared with CD56bright NKLR cells. TIM-3 and KLRG1 were preferentially expressed by both intrahepatic and circulating CD56dim NK cells (Figure 11B). The coexpression of different ICs was evaluated by multiparametric PhenoGraph algorithms (Figure 11, C and D), which generated 3 clusters of CD56bright NK cells (c1–2, c6) and 4 clusters of CD56dim NK cells (c3–5, c7). These analyses confirmed the differential and peculiar NK cell expression patterns of ICs in CRLM. Indeed, CD56bright NKLR cells in c1 and c2 showed higher expression of TIGIT and CD39, each mutually excluded (Figure 11D). High expression of NKG2A was found in c6 embedded in both circulating and intrahepatic CD56bright cells. The 2 CD56dim NKPB cell clusters c4–5, highly enriched in CRLM, were also characterized by high expression of TIM-3, LAIR-1, and KLRG1 (Figure 11D).
 Figure 11
Figure 11Flow cytometry analysis of blood and tissue NK cells in CRLM patients. (A and B) Flow cytometry of immune checkpoint expression in NK cells from HDPB, CRLMPB, and CRLMLR. Data are shown as mean ± SEM. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. One-way ANOVA (left) or 2-way ANOVA (right); n = 10 per group. (C) UMAP projection showing relative abundance of NK cell clusters in HDPB, CRLMPB, and CRLMLR. (D) Dot plot of marker frequency (percent) and MFI across NK cell clusters.
We then evaluated the expression of CXCR4 and IL-1R8 (SIGIRR). IL-1R8 is an IC whose expression is upregulated following NK cell maturation (31, 52). Accordingly, CD56dim NKPB cells expressed high levels of this molecule (Figure 12A). Surprisingly, we also observed an increased amount of IL-1R8 on CD56bright NKPB cells from CRLM patients compared with HDs. In line with our transcriptional scRNA-Seq analysis, both intrahepatic CD56bright and CD56dim NK cells showed the highest levels of IL-1R8 in CRLM (Figure 12A). Similarly, the highest percentages of CXCR4+ cells were found in both intrahepatic CD56bright and CD56dim NK cells (Figure 12, B and C). We then investigated the association between the levels of CXCR4 expression and IFN-γ production. To this end, CD56bright and CD56dim cells were stratified on the basis of the different intensity of CXCR4 expression. First, our results confirmed that CD56bright NKLR cells produced IFN-γ (Figure 12B). Then, CXCR4dim/neg showed higher IFN-γ levels compared with CXCR4bright in CRLM from both intrahepatic CD56bright and CD56dim NK cells (Figure 12, B and C). These results were also confirmed by multiparametric PhenoGraph analysis (Figure 12, D and E). CXCR4 was associated with the liver residency marker CXCR6, TIGIT, and CD39 in cluster 6 (Figure 12E). Finally, CXCR4+CXCR6+ NK cells (cluster 5) were observed only in the PB of patients with CRLM, not in HDs.
 Figure 12
Figure 12Evaluation of IL-1R8 and CXCR4+ NK cell profiling in CRLM. (A) IL-1R8 expression in NK cells from HDPB, CRLMPB, and CRLMLR. Top: Relative frequency (percent) in total NK, CD56bright, and CD56dim subsets. Bottom: The MFI in the same subsets. (B and C) CXCR4 (left) and IFN-γ (right) expression in CD56bright (B) and CD56dim (C) NK cells from HDPB, CRLMPB, and CRLMLR, stratified by CXCR4 levels. (A–C) Data are shown as mean ± SEM. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. One-way ANOVA (A; B and C, left) or 2-way ANOVA (B and C, right). (A) n = 15 (HDPB, CRLMPB), n = 14 (CRLMLR); (B and C) n = 9. (D) UMAP projection showing NK cell cluster distribution across HDPB, CRLMPB, and CRLMLR. (E) Dot plot of marker frequency (percent) and MFI in NK cell clusters.
CXCR4 and IL-1R8 blockade enhances NK cell effector function in CRLM. We next investigated the functional relevance of IL-1R8 and CXCR4 in NKLR and NKPB cells. To properly activate NK cells, we stimulated isolated CRLM lymphocytes and PBMCs with the cytokines IL-2, IL-12, and IL-18 (53). Targeting IL-1R8 with a blocking mAb increased the effector functions of intrahepatic CD56dim NK cells, as indicated by the higher intracellular levels of TNF-α and IFN-γ upon stimulation with IL-2, IL-12, and IL-18, in 5 of 6 and 4 of 6 CRLM patients, respectively (Figure 13, A and B, and Supplemental Figure 8, A and B). These results indicate that despite NK cell subsets expressing comparable levels of IL-1R8, they exhibit distinct functional properties upon IL-1R8 blockade in this experimental condition. In addition, blocking IL-1R8 would result in unleashing specific functional features, depending on the effector potential, transcriptional profile, and repertoire of activating and inhibitory molecules of NK cell subsets (Figure 13C).
 Figure 13
Figure 13Targeting IL-1R8 and CXCR4 in NK cells from CRLM. (A and B) TNF-α (A) and IFN-γ (B) expression in liver CD56dim and CD56bright NK cells from CRLMLR patients stimulated with anti–IL-1R8 mAb or isotype control. (C) Cytokine-stimulated PB NK cells (n = 6, HDs) show increased cytotoxicity upon IL-1R8 blockade in HT-29 killing assay at indicated effector-to-target (E:T) ratios (1:5, 1:2.5, 1:1). (D) IFN-γ expression in CXCR4+CD56dim liver NK cells from CRLM patients stimulated with CXCL12 and/or plerixafor. (A–D) Data are shown as mean ± SEM. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. Statistical analysis was performed using paired 2 tailed parametric t test (A and B), paired 2 tailed non-parametric test (C), and 2-way ANOVA (D). Sample size: n = 3–6 for A–C; n = 4–6 for D.
Finally, we investigated the role of CXCR4 in intrahepatic NK cells by stimulating them with CXCL12 and/or with the CXCR4 antagonist plerixafor. Intrahepatic CD56dim NK cells (but not CD56bright) showed a significantly decreased production of IFN-γ upon stimulation with CXCL12 when compared with controls (Figure 13D). This impairment was restored by plerixafor, which counteracted CXCL12-mediated inhibition of IFN-γ production of intrahepatic CD56dim NK cells (Figure 13D). These results indicate that the effector functions of CXCR4+CD56dim NK cells infiltrating CRLM are tuned by the CXCR4-ligand interactions.
-
Discussion
Heterogeneity of tumor-infiltrating immune cells represents a major challenge in achieving clinical benefits from immunotherapy in CRLM. Several lines of evidence demonstrated that liver tumor microenvironment (TME) actively participates in the establishment and progression of metastases (54, 55). The present study investigates the complexity of matched intrahepatic and circulating NK cells at single-cell level in patients undergoing surgical resections of CRLM lesions after the administration of na-CHT treatment.
CD56bright NK cells are considered precursors of mature CD56dim NK cells (56). In line with other studies (33, 57), we confirmed a developmental trajectory from CD56bright to CD56dim and the existence of transitional differentiation stages in circulating NK cells. The expression of CX3CR1, a marker associated with the tissue recruitment of mature NK cells (40–42), strongly suggests that circulating CD56dim NK cells infiltrate the CRLM and work in synergy with NKLR cells. Conversely, we show here that intrahepatic CD56bright and CD56dim cells in CRLM have different developmental origin, end-terminal differentiation programs, and conserved effector gene signatures. The CD56bright cell subsets represented the vast majority of NKLR cells and expressed liver-residency genes (CXCR6, CCR5, EOMES, and CD69). Moreover, they are characterized by TFs such as IRF8, EOMES, and TOX2 that are required for functional maturation of human NK cells (35–37, 39). For instance, TOX2 upregulation is directly involved in maturation and cytotoxicity in human NK cells (35). In agreement with TF profiling, intrahepatic CD56bright NK cells from CRLM patients expressed high levels of effector function molecules (e.g., GZMK, GZMA, FASL) (43, 58, 59). Hence, in contrast with their circulating counterparts, CD56bright NKLR cells can be considered mature and terminally differentiated lymphocytes. Moreover, tissue-resident NK cells expressed cluster markers of CD56dim cells from PB (e.g. CCL4, CCL5, NKG7, and IL32) (33), reflecting the unique ontogeny and activation state in the CRLM contexture. Single-cell transcriptomic profiling revealed a strong similarity between NK cells isolated from PT and IM tissues, suggesting that NK cells undergo early commitment and functional adaptation independently of direct tumor–NK cell interactions. This highlights the potential role of soluble factors in modulating NK cell differentiation and antitumor activity. In contrast, myeloid cell clusters from the same samples displayed distinct distributions between the two regions, indicating a more localized TME-dependent adaptation (48).
We recently identified a subset of tumor-infiltrating γδ T lymphocytes that recirculate in PB of CRLM patients, where they retained a transcriptional signature and γδ T cell receptor repertoire resembling their liver origin and positively correlated with longer overall survival in CRLM patients (60). Along this line, we detected rare circulating NK cells showing liver signatures, suggesting that intrahepatic NK cells leave CRLM and recirculate into the bloodstream, where they retain distinctive transcriptional profiles, but further studies are needed to evaluate their association with the clinical outcome.
Clinical trials are investigating the outcome of CRLM patients treated with immune checkpoint inhibitors (ICIs) (61, 62). Hence, the characterization of IC pattern among tumor-infiltrating NK cells is key to optimize immunotherapies. The present study depicts specific expression patterns of ICs by CD56bright and CD56dim NK cells in CRLM patients. CD56bright cells expressed high levels of NKG2A, which is currently under assessment in several clinical trials (63, 64). Higher levels of TIM-3, KLRG1, and KIRs were found in CD56dim NK cells, whose targeting is under investigation (62). In addition, we identified TIGIT, CD96, and CD39 as ICs specifically expressed by CD56bright NKLR cells. Notably, TIGIT and CD39 were shown to be upregulated in PB NK cells from patients with acute myeloid leukemia and in esophageal squamous cell carcinoma, respectively (65, 66), suggesting specific IC regulation mechanisms that depend on the tumor type.
The clinical relevance of NK cell–mediated immune surveillance in liver metastatic diseases had been extensively reported (22, 29–31). Here, we identified an IFNGhigh signature associated with both intrahepatic CD56dim and CD56bright NK cells from CRLM. IFN-γ is the critical effector factor determining the success of immunotherapy and represents an important prediction marker for the clinical response to ICIs (30). Indeed, high IFNG and IFNG-related gene signatures in patients with different primary and metastatic tumors (i.e., melanoma, head and neck squamous cell carcinoma, gastric cancer, lung cancer) were associated with effective ICI treatments (67–70). Recent studies showed that IFN-γ–secreting lymphocytes modify the TME (71, 72). IFN-γ administration has been proposed to improve the efficacy of PD-1 blockade therapy in pancreatic cancer (73) and is under clinical evaluation (e.g., ClinicalTrials.gov NCT02614456, NCT03063632). The IFNGhigh signature identified in our study is also enriched in several activation molecules (JUN, GIMAP4, DUSP), suggesting the antitumor potentiality of liver NK cells. On the other hand, the signature is enriched also in stress-induced molecules (HSPs), which have been associated with a dysfunctional state (57).
In this study, we show that IL-1R8 is ubiquitously and highly expressed by all human intrahepatic NK cells in CRLM. We have previously shown that IL-1R8 acts as an IC dampening IL-18–dependent NK cell activation in mouse models of primary and metastatic liver cancer (31). Here we show that myeloid-derived IL-18 emerges as a pivotal upstream regulator in CRLMs. We also provide evidence that an anti–IL-1R8 blocking mAb increases the functional activation of cytokine-stimulated intrahepatic NK cells, although with variability among patients, supporting the hypothesis that IL-1R8 could be a target for immunotherapy. In preclinical models, IL-1R8 deficiency increased susceptibility to colitis-associated chemical carcinogenesis (74), while enhancing protection from primary liver carcinogenesis and CRLM (23). These opposite effects are likely explained by the fact that IL-1R8 is expressed in colon epithelial cells as well as NK cells. IL-1R8 systemic blockade has the risks associated with exacerbated inflammatory reactions, in parallel with unleashed antitumor activity of NKLR cells. The development of IL-1R8 as IC in cancer would imply NK-directed targeting, for instance through genetic silencing.
In addition, we observed that both intrahepatic CD56dim and CD56bright NK cell subsets displaying IFNGhigh signatures were further characterized by lower expression of CXCR4, a chemokine receptor involved in leukocyte bone marrow retention and in NK cell maturation (75, 76). Recent studies extended its biological significance, showing that hepatic stellate cells and fibroblasts secrete CXCL12 (11), which, in turn, hampered NK cell–mediated resistance against liver metastasis by engaging CXCR4 (44). We observed that among CD45+ liver cells, KCs expressed high levels of CXCL12, and that the stimulation of intrahepatic CD56dim NK cells with CXCL12 limited their functional activity, which was rescued by the CXCR4 antagonist plerixafor.
In summary, the present study discloses the origin, heterogeneity, and effector functions of matched circulating and intrahepatic NK cells from CRLM patients by using single-cell technologies and unbiased analytic approaches, shedding light on an abundant IFNGhigh resident CD56bright NK cell population with an antitumor signature, and providing two possible therapeutic targets (IL-1R8 and CXCR4) to complement current immunotherapy of CRLM.
-
Methods
Sex as a biological variable
Our study examined male and female individuals, and sex was not considered as a key biological variable.
Study design
The study investigated the role of NK cells in patients with histologically proven CRLM older than 18 years of age who underwent hepatectomy at IRCCS (Istituto di Ricovero e Cura a Carattere Scientifico [Scientific Institute for Research, Hospitalization and Healthcare]) Humanitas Research Hospital, mostly represented by patients with synchronous clinical profile treated with standard combination bevacizumab/cetuximab na-CHT 2–4 weeks before surgery. The preoperative work-up consisted of liver-specific MRI and total-body contrast-enhanced CT, performed a maximum of 30 days before surgery. Matched specimens from the invasive margin (IM) and peritumor (PT) area as well as from PBMCs of 3 patients with CRLM were processed for scRNA-Seq. Specifically, scRNA-Seq analysis was performed on flow cytometry–sorted NK cells (Lin–CD56+) excluding CD3+ lymphocytes and myeloid and B cells (altogether referred to as Lin+), and on total CD45+ leukocytes that were freshly isolated from the matched samples of 3 representative patients (2 male and 1 female) as well as on matched PBMCs.
Tissue processing and cell preparations
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats of healthy donors (HDs) or from PB of CRLM patients by Lympholyte Cell Separation density gradient solution (Cedarlane Laboratories) according to the manufacturer’s instructions. Liver tissues were dissociated by enzymatic digestion in Hanks balanced salt solution with Ca2+ and Mg2+ (HBSS+/+, Euroclone SpA) with 2 mg/mL of collagenase D (Sigma-Aldrich), 50 μg/mL DNase I (Sigma-Aldrich), 2% fetal bovine serum (FBS) (Sigma-Aldrich), and 10 mmol/L HEPES (Lonza) in gentleMACS Dissociator (Miltenyi) for 35 minutes at 37°C/5% CO2. Cells were then filtered through a 70 μm cell strainer (Corning) and washed with HBSS without Ca2+ and Mg2+ (Euroclone SpA). Lymphocytes were separated by 70%/30% discontinuous Percoll gradient (Cytiva) and frozen in FBS (Lonza) with 10% of dimethylsulfoxide (Lonza) before being stored in liquid nitrogen for further analysis.
Flow cytometry analysis
For multiparametric flow cytometry analysis, cells were stained with Zombie Aqua fixable viability kit (BioLegend) for live/dead discrimination. Then, cells were washed with HBSS–/– with 2% of FBS (FACS buffer) and incubated with the mix of mAbs for 20 minutes in the dark at room temperature, washed again with FACS buffer, and fixed in 1% of paraformaldehyde (Santa Cruz Biotechnology). All reagents and mAbs are listed in Supplemental Table 1.
The intracellular concentrations of IFN-γ and TNF-α were measured by flow cytometry using anti–human IFN-γ mAb (clone B27; BioLegend, catalog 506518) and anti–human TNF-α mAb (clone Mab11; BioLegend, catalog 506518) and were evaluated after stimulation of PBMCs and liver-infiltrating lymphocytes with 1 μg/mL of GolgiPlug protein transport inhibitor (BD Biosciences). After 4 hours at 37°C/5% CO2, cells were collected and washed with HBSS–/–, and cellular membrane staining was done as described above. Subsequently, intracellular staining was performed with a Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s instructions.
All samples were acquired by a FACSymphony A5 flow cytometer (BD Biosciences). Flow cytometry data were compensated using single-stained controls with BD Compbeads (BD Biosciences) conjugated to the specific fluorescent mAb. For accurate flow cytometry practice, all mAbs were previously titrated (77). All flow cytometry data, including the dimensionality reduction method with uniform manifold approximation and projection (UMAP) algorithm and unsupervised clustering with PhenoGraph algorithm, were analyzed by FlowJo software version 10.8.1 (FlowJo LLC).
Library preparation and sample sequencing
NK cells were sorted by flow cytometry (BD FACSAria III Cell Sorter) as lineage negative and CD56+, isolated from PB and liver of 3 CRLM patients, and were subjected to scRNA-Seq analysis. NK cell purity after FACS sorting was 97% or greater. In collaboration with the Genomic Unit of Humanitas Research Hospital, scRNA-Seq libraries were prepared using the Single Cell 3′ Library and Gel Bead Kits (10x Genomics). According to the established protocol, approximately 10,000 cells for each sample were loaded into the Chromium controller (10x Genomics) to generate the Gel Beads-in-Emulsion (GEMs). The libraries were sequenced using the Illumina NextSeq500 platform.
Processing scRNA-Seq data. 3′-based sequencing data were aligned and quantified using the Cell Ranger Single-Cell Software Suite (v3.0.2, 10x Genomics) to the GRCh38 (refdata-gex-GRCh38-2020-A) human reference genome. Subsequent analyses were performed using the R package Seurat (78) (v3.1.1; R v3.6.1). Cells were filtered based on quality metrics to exclude low-quality, potentially dying cells, or doublets. Specifically, cells expressing fewer than 200 genes or more than 6,400 genes, with fewer than 365 unique molecular identifiers (UMIs) or more than 43,000 UMIs, or with mitochondrial gene content exceeding 20% were removed. PB NK cells and integrated PT and IM NK cells from 3 CRLM patients were first analyzed separately. Specifically, the FindIntegrationAnchors() and IntegrateData() functions were used to combine tissue NK cells of 3 CRLM patients as well as NK cells from PB. Principal component analysis (PCA) was performed with the RunPCA() function. To reduce the dimensionality of the dataset, the RunUMAP() function was used on the pre-computed principal components. The clusters were identified with the FindClusters() function based on pre-computed principal components and visualized by UMAP plot. Eighteen and fourteen clusters were identified at resolution 0.7 for tissue and PB, respectively. PT and PB NK cells were then integrated, and 18 clusters were identified at resolution parameter 0.8. For each integrated dataset, NK cell clusters were annotated based on the average expression of canonical NK cell markers. The default Wilcoxon’s rank-sum test was used by running of the FindAllMarkers() function to find differentially expressed markers in each cluster. The upregulated or downregulated genes with |log2(fold change)| greater than 0.25 and P value less than 0.05 were considered significant DEGs.
Gene Ontology enrichment analysis
Gene Ontology (GO) (biological process) enrichment analysis was performed by the R package clusterProfiler (79). The GO pathways of each cell type were enriched using DEGs with FDR-adjusted P value ≤ 0.05 and log(fold change) > 0.25. Only enriched GO terms with q value ≤ 0.01 were selected as significant and visualized by the R package ggplot2. Dots were sized by adjusted P value and colored by EnrichFold values defined as the ratio between BgRatio and GeneRatio terms from enrichment analysis. IFNGhigh signature was defined by selection of shared DEGs with FDR-adjusted P value ≤ 0.05 and log(fold change) > 0.25, from the comparison of c5LR versus c0–1–2LR and c6LR versus c3–4LR.
Pseudotime ordering
Monocle2. Pseudotime trajectory analysis was performed separately on tissue and PB by Monocle (v2.8.0) (80). Cells were ordered along the trajectory by the dpFeature unsupervised procedure. Only genes expressed in at least 5% of all the cells were selected, and the top 1,000 highly variable genes (HVGs) were used as the ordering genes. For tissue trajectory, c8LR was selected as the root node of the graph, while for blood trajectory, c8PB was selected as the root node. The DifferentialGeneTest() function was used to test for a significant correlation between gene expression and pseudotime. A gene was defined as significantly associated with pseudotime if its estimated q value was lower than 0.01. Significantly associated genes were visualized by the plot_pseudotime_heatmap() function.
Slingshot. Pseudotime trajectory inference was performed using the Slingshot algorithm (v2.12.0) (81) in R (v4.4.0) and Seurat (v4.4.0). Only cells belonging to NK cell clusters from tissue and blood objects were selected for downstream analysis. On the selected cells, a new UMAP was computed to include only NK cells.
Subsequently, the 2 new datasets were independently converted into a SingleCellExperiment object, using the log-normalized expression matrix and cluster identities. UMAP embeddings were imported into the SingleCellExperiment object using the reducedDim() function. Slingshot was run using the slingshot() function, specifying the original Seurat clusters as lineage labels and the UMAP embedding as the reduced dimensionality space and setting cluster 8 as the starting point for both the tissue and blood. The distance method was set to “simple,” and the omega parameter was enabled to allow adaptive weighting of constraints.
Pseudotime values were extracted using the slingPseudotime() function, and shared pseudotime was calculated as the mean pseudotime across the inferred lineages.
Public healthy liver scRNA-Seq databases
Raw counts of healthy liver samples (n = 5) were downloaded from the NCBI’s Gene Expression Omnibus (GEO) database (GSE136103) (45) and processed in Python (v3.9.7) using Scanpy (v1.9.3) (82). Each sample underwent independent quality control: cells with fewer than 500 or more than 4,000 expressed genes, fewer than 950 or more than 12,000 UMIs, greater than 10% mitochondrial content, or less than 0.05% ribosomal content were filtered out. Doublets were removed using Scrublet (v0.2.3) (83). Samples were merged into a single AnnData object (v0.10.4) (84) and log-normalized (scale factor 10,000). HVGs (n = 4,000) were identified using Seurat_v3 dispersion-based methods, followed by PCA. Batch correction and data integration were performed with pyHarmony (v1.0.21) (85) setting “patient_ID” as batch key. Clustering was performed using the Leiden algorithm (86) and visualized via UMAP (87) in the Harmony-corrected space. For NK cell re-clustering, selected cells were extracted, HVGs and PCA were re-computed, and integration was repeated with the same parameters.
Integration of healthy liver and tumor-associated NK cells
Raw count matrices of tumor-associated NK cells were first exported from Seurat and imported into Python (v3.9.7) for integration with healthy NK cells. Data integration and batch correction were performed using the single-cell Variational Inference (scVI)-tools algorithm (v1.0.0) (88), setting both “sequencing_ID” and “sample_ID” as batch key covariates.
For DEG analysis, the raw count matrix of the integrated dataset was exported from Python and imported into R (v4.4.0) as a Seurat object, generated using the CreateSeuratObject() function (Seurat v4.4.0). DEGs were computed using the Seurat FindMarkers() function, applying a negative binomial test, setting “sequencing_ID” for batch correction. Significant DEGs were defined as those with an adjusted P value less than 0.05 and |log2(fold change)| > 0.25.
NicheNet interaction analysis
The analysis to predict the existing ligand-target interaction between the CD45+ population and NK cells was done with NicheNet (v1.0.0, Seurat v3.6). The normalized data obtained after the Seurat analysis were imputed using the ALRA (adaptively thresholded low-rank approximation) function to correct the missed counts, with a predicted rank-k approximation of 35. We set as sender cells, which express the ligand genes, each cluster in the CD45+ cell population, and as receiver cells 3 different NK cell clusters (CD56bright, CD56dim, and cycling NK cells). The analysis was restricted to the genes expressed in at least 50% of sender and receiver cell clusters. The cluster gene markers with adjusted P value 0.05 and avg_logFC 0.5 from each receiver cell population were used as the target gene set, whereas the background genes were chosen as expressed in all cell populations with a cluster frequency above 50%. After the ligand activity analysis, only the ligands with a positive Pearson’s correlation coefficient were considered. To visualize the ligand-receptor interactions, we used the circlize R package (v0.4.12).
We gathered together ligands related to myeloid or lymphoid compartments and selected the first 100 interactions ranked by their regulatory potential.
In vitro stimulation
Human NK cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 1% l-glutamine, 1% penicillin/streptomycin with IL-2 (Proleukyn), IL-12 (Miltenyi), and IL-18 (MBL Life Science). The following reagents were used, as specified: an original mouse anti-hSIGIRR antibody (IgG1, κ isotype, low endotoxin, azide free; generated by the University of Rijeka Faculty of Medicine, Center for Proteomics); Ultra-LEAF Purified Mouse IgG1, κ Isotype Control (Ctrl) Antibody (BioLegend); CXCL12 (PeproTech); plerixafor; and Cell Activation Cocktail (eBioscience), IFN-γ and TNF-α production was evaluated after 16 hours of stimulation with IL-2 (10 ng/mL), IL-12 (10 ng/mL), IL-18 (5 ng/mL), anti-hSIGIRR (1 μg/mL), Ultra-LEAF Purified Mouse IgG1, κ Isotype Ctrl Antibody (1 μg/mL), CXCL12 (100 ng/mL), and plerixafor (100 ng/mL) alone or in combination. The intracellular staining was performed using a BD Cytofix/Cytoperm Fixation/Permeabilization Kit following the manufacturer’s instructions. BD GolgiPlug (containing brefeldin) was added 4 hours before intracellular staining.
Killing assay
For cytotoxicity experiments, PBMCs were isolated from buffy coats by Lympholyte Cell Separation density gradient solution (Cedarlane Laboratories) according to the manufacturer’s instructions. NK cells were purified by negative selection, using EasySep Human NK Cell Enrichment Kit (Stemcell Technologies), and cultured for 6 days in DMEM/F-12–DMEM high-glucose (1:1) supplemented with 10% Human Serum, Type AB (Capricorn Scientific); 1% l-glutamine; 1% penicillin/streptomycin; 1% sodium pyruvate; 1% non-essential amino acids; IL-15 (10 ng/mL; Miltenyi); and IL-2 (100 IU/mL; Proleukyn). On the day of coculture, HT-29 target cells were detached with Trypsin/EDTA solution (Lonza), washed twice in PBS, and labeled with CellTrace CFSE Cell Proliferation Kit (Thermo Fisher Scientific) following the manufacturer’s protocol. Fifty thousand labeled HT-29 cells were seeded in a flat-bottom 96-well plate 4 hours before the coculture. Cytokine-primed NK cells were collected and cocultured with HT-29 cells at different effector-to-target ratios (1:5, 1:2.5, 1:1) upon 30 minutes of preincubation with anti–hIL-1R8 (1 μg/mL) or Ultra-LEAF Purified Mouse IgG1, κ Isotype Ctrl Antibody (1 μg/mL) at 37°C, which was left throughout the duration of the coculture. Cells were stimulated with IL-15 (20 ng/mL; Miltenyi) and IL-18 (50 ng/mL; MBL Life Science) overnight. To quantify viable HT-29 target cells, supernatants were collected and pooled with the remaining adherent HT-29 cells detached using the trypsin. The number was normalized using CountBright Absolute Counting Beads (Thermo Fisher Scientific). Killing activity was calculated as follows: 1 – (residual number of target cells in the presence of NK cells / number of control target cells).
Statistics
Flow cytometry analysis was performed using GraphPad Prism v9. ANOVA with Tukey’s multiple-comparison test was used to compare multiple groups. The data are presented as median value ± SEM. Statistically significant P values are represented in GraphPad style and are summarized with asterisks as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Study approval
The study protocol was in accordance with the ethical guidelines established in the Declaration of Helsinki, and participants gave written informed consent to participate in the study before taking part. All patients were recruited at the Hepatobiliary and General Surgery Department at Humanitas Clinical and Research Center (HCR), Rozzano, Milan, Italy. Liver tissue and PB specimens were collected from CRLM patients in accordance with clinical protocols approved by the Institutional Review Board of HCR Institute (approval 168/18). PB of HDs was obtained in accordance with HCR and with clinical protocols approved by the Institutional Review Board of Desio Hospital, Milan, Italy.
Data availability
All data relevant to the study are included in the article or its supplemental information. The filtered scRNA-Seq gene expression counts generated in this study are publicly available in the Zenodo Repository with accession number 10.5281/zenodo.17366198. The raw counts are publicly available in the Sequence Read Archive database ID SRP635148 under the BioProject ID PRJNA1346442. The accession code for the CD45+ dataset repository in GEO is GSE200253. Values for all data points in graphs are reported in the Supporting Data Values file.
-
Author contributions
MD, GC, and GT were responsible for patient recruitment and collection of biological specimens and contributed to the study design. DS, VC, EB, RG, SC, JZ, and FS processed samples and conducted experiments. PM, ST, RC, RP, PK, AD, and SNM analyzed the scRNA-Seq data and interpreted the results. CP designed and performed the scRNA-Seq sample preparation. JM, DS, EM, AM, CG, and DM interpreted the results. JM, DS, CG, and DM designed and conducted the study and wrote the manuscript. CG and DM directed and supervised the study. JM and DS share first authorship, having contributed equally in conceiving the overall study. The order of the first authors reflects the leadership exerted in the study.
-
Funding support
- Italian Association for Cancer Research (AIRC) (AIRC 5x1000 project 21147 to AM; Fellowships for Italy Post-Doc or PhD to DS, SC, and VC).
- European Union – Next Generation EU (PNRR-MAD-2022-12375947 and PNC-E3-2022-23683266 PNC-HLS-DA to CG).
- Italian Ministry of University and Research (PRIN 20174T7NXL and 2022NKEXAT to CG).
- Competitive fellowships awarded from the PhD program of Experimental Medicine at the University of Milan to PM.
- A competitive fellowship awarded from the Data Science in Medicine and Nutrition (DASMEN) PhD program at Humanitas University to ST.
- Purchase of a FACSymphony A5 was defrayed in part by a grant from the Italian Ministry of Health (Agreement 82/2015).
- The publication fee for this work was covered by the Italian Ministry of Health’s “Ricerca Corrente” funding to the IRCCS Humanitas Research Hospital
-
Acknowledgments
The authors express their gratitude to all patients and healthy donors for their generous participation in this study, as well as to the nurses and clinicians at IRCCS Humanitas Research Hospital, Milan, Italy, for their invaluable assistance. Special thanks to Federico S. Colombo and the entire Flow Cytometry facility at IRCCS Humanitas Research Hospital for their technical expertise and support.
Address correspondence to: Cecilia Garlanda, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Email: cecilia.garlanda@humanitasresearch.it. Or to: Domenico Mavilio, Unit of Clinical and Experimental Immunology, Department of Medical Biotechnology and Translational Medicine, IRCCS Humanitas Research Hospital, Via Alessandro Manzoni, 56, 20089 Rozzano, Milan, Italy. Phone: 39.02.8224.5157; Email: domenico.mavilio@unimi.it.
-
Footnotes
Conflict of interest: CG and AM are inventors on a patent related to IL-1R8 (WO2019081591).
Copyright: © 2025, Mikulak et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2025;135(24):e190778.https://doi.org/10.1172/JCI190778.
-
References
- Tsilimigras DI, et al. Liver metastases. Nat Rev Dis Primers. 2021;7(1):27.
- Reboux N, et al. Incidence and survival in synchronous and metachronous liver metastases from colorectal cancer. JAMA Netw Open. 2022;5(10):e2236666.
- Guo M, et al. Neoadjuvant chemotherapy for colorectal liver metastases: a contemporary review of the literature. World J Gastrointest Oncol. 2021;13(9):1043–1061.
- Famularo S, et al. Hepatectomy versus chemotherapy for resectable colorectal liver metastases in progression after perioperative chemotherapy: expanding the boundaries of the curative intent. Cancers (Basel). 2023;15(3):783.
- Yu X, et al. Emerging role of immunotherapy for colorectal cancer with liver metastasis. Onco Targets Ther. 2020;13:11645–11658.
- Waldman AD, et al. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. 2020;20(11):651–668.
- Bilen MA, et al. Sites of metastasis and association with clinical outcome in advanced stage cancer patients treated with immunotherapy. BMC Cancer. 2019;19(1):857.
- Botticelli A, et al. The agnostic role of site of metastasis in predicting outcomes in cancer patients treated with immunotherapy. Vaccines (Basel). 2020;8(2):203.
- Dai Y, et al. Perspectives on immunotherapy of metastatic colorectal cancer. Front Oncol. 2021;11:659964.
- Wei XL, et al. PD-L1 expression in liver metastasis: its clinical significance and discordance with primary tumor in colorectal cancer. J Transl Med. 2020;18(1):475.
- Xue R, et al. Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature. 2022;612(7938):141–147.
- Zhao M, et al. Current insights into the hepatic microenvironment and advances in immunotherapy for hepatocellular carcinoma. Front Immunol. 2023;14:1188277.
- Marmorino F, et al. Immune checkpoint inhibitors in pMMR metastatic colorectal cancer: a tough challenge. Cancers (Basel). 2020;12(8):2317.
- Nazemalhosseini Mojarad E, et al. Low level of microsatellite instability correlates with poor clinical prognosis in stage II colorectal cancer patients. J Oncol. 2016;2016:2196703. View this article via: PubMed Google Scholar
- Wang Y, et al. Radiomics and radiogenomics in evaluation of colorectal cancer liver metastasis. Front Oncol. 2021;11:689509.
- Malki A, et al. Molecular mechanisms of colon cancer progression and metastasis: recent insights and advancements. Int J Mol Sci. 2020;22(1):130.
- Cohen R, et al. Association of primary resistance to immune checkpoint inhibitors in metastatic colorectal cancer with misdiagnosis of microsatellite instability or mismatch repair deficiency status. JAMA Oncol. 2019;5(4):551–555.
- Andre T, et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383(23):2207–2218.
- Zhao J, et al. Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. 2020;6:22.
- Cazzetta V, et al. Natural killer-dendritic cell interactions in liver cancer: implications for immunotherapy. Cancers (Basel). 2021;13(9):2184.
- Mikulak J, et al. Hepatic natural killer cells: organ-specific sentinels of liver immune homeostasis and physiopathology. Front Immunol. 2019;10:946.
- Polidoro MA, et al. Tumor microenvironment in primary liver tumors: a challenging role of natural killer cells. World J Gastroenterol. 2020;26(33):4900–4918.
- Vivier E, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44–49.
- Marcenaro E, et al. Editorial: NK cell subsets in health and disease: new developments. Front Immunol. 2017;8:1363.
- Scoville SD, et al. Modeling human natural killer cell development in the era of innate lymphoid cells. Front Immunol. 2017;8:360.
- Hudspeth K, et al. Human liver-resident CD56(bright)/CD16(neg) NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways. J Autoimmun. 2016;66:40–50.
- Stegmann KA, et al. CXCR6 marks a novel subset of T-bet(lo)Eomes(hi) natural killer cells residing in human liver. Sci Rep. 2016;6:26157.
- Hesslein DG, Lanier LL. Transcriptional control of natural killer cell development and function. Adv Immunol. 2011;109:45–85.
- Ducimetiere L, et al. Conventional NK cells and tissue-resident ILC1s join forces to control liver metastasis. Proc Natl Acad Sci U S A. 2021;118(27):e2026271118.
- Donadon M, et al. Increased infiltration of natural killer and T cells in colorectal liver metastases improves patient overall survival. J Gastrointest Surg. 2017;21(8):1226–1236.
- Molgora M, et al. IL-1R8 is a checkpoint in NK cells regulating anti-tumour and anti-viral activity. Nature. 2017;551(7678):110–114.
- Mikulak J, et al. NKp46-expressing human gut-resident intraepithelial Vδ1 T cell subpopulation exhibits high antitumor activity against colorectal cancer. JCI Insight. 2019;4(24):e125884.
- Rebuffet L, et al. High-dimensional single-cell analysis of human natural killer cell heterogeneity. Nat Immunol. 2024;25(8):1474–1488.
- Crinier A, et al. Single-cell profiling reveals the trajectories of natural killer cell differentiation in bone marrow and a stress signature induced by acute myeloid leukemia. Cell Mol Immunol. 2021;18(5):1290–1304.
- Vong QP, et al. TOX2 regulates human natural killer cell development by controlling T-BET expression. Blood. 2014;124(26):3905–3913.
- Mace EM, et al. Biallelic mutations in IRF8 impair human NK cell maturation and function. J Clin Invest. 2017;127(1):306–320.
- Adams NM, et al. Transcription factor IRF8 orchestrates the adaptive natural killer cell response. Immunity. 2018;48(6):1172–1182.
- Jeevan-Raj B, et al. The transcription factor Tcf1 contributes to normal NK cell development and function by limiting the expression of granzymes. Cell Rep. 2017;20(3):613–626.
- Bi J, Wang X. Molecular regulation of NK cell maturation. Front Immunol. 2020;11:1945.
- Ran GH, et al. Natural killer cell homing and trafficking in tissues and tumors: from biology to application. Signal Transduct Target Ther. 2022;7(1):205.
- Hertwig L, et al. CX3CR1-dependent recruitment of mature NK cells into the central nervous system contributes to control autoimmune neuroinflammation. Eur J Immunol. 2016;46(8):1984–1996.
- Fraticelli P, et al. Fractalkine (CX3CL1) as an amplification circuit of polarized Th1 responses. J Clin Invest. 2001;107(9):1173–1181.
- Yang C, et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat Commun. 2019;10(1):3931.
- Correia AL, et al. Hepatic stellate cells suppress NK cell-sustained breast cancer dormancy. Nature. 2021;594(7864):566–571.
- Ramachandran P, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575(7783):512–518.
- Sparano C, et al. Autocrine TGF-β1 drives tissue-specific differentiation and function of resident NK cells. J Exp Med. 2025;222(3):e20240930.
- Browaeys R, et al. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020;17(2):159–162.
- Cortese N, et al. High-resolution analysis of mononuclear phagocytes reveals GPNMB as a prognostic marker in human colorectal liver metastasis. Cancer Immunol Res. 2023;11(4):405–420.
- Bodzioch M, et al. Evidence for potential functionality of nuclearly-encoded humanin isoforms. Genomics. 2009;94(4):247–256.
- Zuccato CF, et al. Mitochondrial-derived peptide humanin as therapeutic target in cancer and degenerative diseases. Expert Opin Ther Targets. 2019;23(2):117–126.
- Schlums H, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. 2015;42(3):443–456.
- Sarhan D, et al. Adaptive NK cells resist regulatory T-cell suppression driven by IL37. Cancer Immunol Res. 2018;6(7):766–775.
- Konjevic GM, et al. The role of cytokines in the regulation of NK cells in the tumor environment. Cytokine. 2019;117:30–40.
- Tang J, et al. Single-cell exome sequencing reveals multiple subclones in metastatic colorectal carcinoma. Genome Med. 2021;13(1):148.
- Hartung F, et al. A core program of gene expression characterizes cancer metastases. Oncotarget. 2017;8(60):102161–102175.
- Di Vito C, et al. NK cells to cure cancer. Semin Immunol. 2019;41:101272.
- Netskar H, et al. Pan-cancer profiling of tumor-infiltrating natural killer cells through transcriptional reference mapping. Nat Immunol. 2024;25(8):1445–1459.
- Smith SL, et al. Diversity of peripheral blood human NK cells identified by single-cell RNA sequencing. Blood Adv. 2020;4(7):1388–1406.
- Wagner JA, et al. CD56bright NK cells exhibit potent antitumor responses following IL-15 priming. J Clin Invest. 2017;127(11):4042–4058.
- Bruni E, et al. Intrahepatic CD69+Vδ1 T cells re-circulate in the blood of patients with metastatic colorectal cancer and limit tumor progression. J Immunother Cancer. 2022;10(7):e004579.
- Shek D, et al. Immune-checkpoint inhibitors for metastatic colorectal cancer: a systematic review of clinical outcomes. Cancers (Basel). 2021;13(17):4345.
- Khan M, et al. NK cell-based immune checkpoint inhibition. Front Immunol. 2020;11:167.
- Fisher JG, et al. Disruption of the NKG2A:HLA-E immune checkpoint axis to enhance NK cell activation against cancer. Vaccines (Basel). 2022;10(12):1993.
- Cazzetta V, et al. NKG2A immune checkpoint in Vδ2 T cells: emerging application in cancer immunotherapy. Cancers (Basel). 2023;15(4):1264.
- Liu G, et al. Increased TIGIT expressing NK cells with dysfunctional phenotype in AML patients correlated with poor prognosis. Cancer Immunol Immunother. 2022;71(2):277–287.
- Zheng Y, et al. IL-6-induced CD39 expression on tumor-infiltrating NK cells predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Immunol Immunother. 2020;69(11):2371–2380.
- Ayers M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940.
- Higgs BW, et al. Interferon gamma messenger RNA signature in tumor biopsies predicts outcomes in patients with non-small cell lung carcinoma or urothelial cancer treated with durvalumab. Clin Cancer Res. 2018;24(16):3857–3866.
- Karachaliou N, et al. Interferon gamma, an important marker of response to immune checkpoint blockade in non-small cell lung cancer and melanoma patients. Ther Adv Med Oncol. 2018;10:1758834017749748.
- Gao J, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167(2):397–404.
- Thibaut R, et al. Bystander IFN-γ activity promotes widespread and sustained cytokine signaling altering the tumor microenvironment. Nat Cancer. 2020;1(3):302–314.
- Hoekstra ME, et al. Long-distance modulation of bystander tumor cells by CD8+ T cell-secreted IFNγ. Nat Cancer. 2020;1(3):291–301.
- Zhang M, et al. Interferon gamma inhibits CXCL8-CXCR2 axis mediated tumor-associated macrophages tumor trafficking and enhances anti-PD1 efficacy in pancreatic cancer. J Immunother Cancer. 2020;8(1):e000308.
- Garlanda C, et al. Increased susceptibility to colitis-associated cancer of mice lacking TIR8, an inhibitory member of the interleukin-1 receptor family. Cancer Res. 2007;67(13):6017–6021.
- Noda M, et al. CXCL12-CXCR4 chemokine signaling is essential for NK-cell development in adult mice. Blood. 2011;117(2):451–458.
- Mayol K, et al. Sequential desensitization of CXCR4 and S1P5 controls natural killer cell trafficking. Blood. 2011;118(18):4863–4871.
- Brummelman J, et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8+ T cells infiltrating human tumors. J Exp Med. 2018;215(10):2520–2535.
- Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.
- Yu G, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287.
- Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386.
- Street K, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19(1):477.
- Wolf FA, et al. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):15.
- Wolock SL, et al. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8(4):281–291.
- Virshup I, et al. anndata: Access and store annotated data matrices. J Open Source Softw. 2024;9(101):4371. View this article via: CrossRef Google Scholar
- Korsunsky I, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–1296.
- Traag VA, et al. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep. 2019;9(1):5233.
- Becht E, et al. Dimensionality reduction for visualizing single-cell data using UMAP. [published online December 3, 2018]. Nat Biotechnol. https://doi:10.1038/nbt.4314. View this article via: PubMed Google Scholar
- Gayoso A, et al. A Python library for probabilistic analysis of single-cell omics data. Nat Biotechnol. 2022;40(2):163–166.
-
Version history
- Version 1 (October 28, 2025): In-Press Preview
- Version 2 (December 15, 2025): Electronic publication



Copyright © 2026 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)