Advertisement
Research ArticleHematologyInflammationOncology
Open Access |  10.1172/JCI184021
10.1172/JCI184021
TP53 mutations and TET2 deficiency cooperate to drive leukemogenesis and establish an immunosuppressive environment
Pu Zhang,1,2,3 Ethan C. Whipp,1 Sarah J. Skuli,4 Mehdi Gharghabi,1 Caner Saygin,5 Steven A. Sher,1 Martin Carroll,4 Xiangyu Pan,6 Eric D. Eisenmann,3 Tzung-Huei Lai,1 Bonnie K. Harrington,7 Wing Keung Chan,1 Youssef Youssef,1 Bingyi Chen,2 Alex Penson,2 Alexander M. Lewis,2 Cynthia R. Castro,2 Nina Fox,2 Ali Cihan,8 Jean-Benoit Le Luduec,2 Susan DeWolf,2 Tierney Kauffman,1 Alice S. Mims,1 Daniel Canfield,1 Hannah Phillips,1 Katie E. Williams,1 Jami Shaffer,1 Arletta Lozanski,1 Tzyy-Jye Doong,1 Gerard Lozanski,1 Charlene Mao,1 Christopher J. Walker,1,9 James S. Blachly,1 Anthony F. Daniyan,8 Lapo Alinari,1 Robert A. Baiocchi,1 Yiping Yang,1 Nicole R. Grieselhuber,1 Moray J. Campbell,10 Sharyn D. Baker,3 Bradley W. Blaser,1 Omar Abdel-Wahab,2 and Rosa Lapalombella1
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Zhang, P. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Whipp, E. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Skuli, S. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Gharghabi, M. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Saygin, C. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by
Sher, S.
in:
PubMed
|
Google Scholar
|

1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by
Carroll, M.
in:
PubMed
|
Google Scholar
|

1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Pan, X. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Eisenmann, E. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Lai, T. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Harrington, B. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Chan, W. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Youssef, Y. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Chen, B. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Penson, A. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Lewis, A. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Castro, C. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Fox, N. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Cihan, A. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Le Luduec, J. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by
DeWolf, S.
in:
PubMed
|
Google Scholar
|

1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Kauffman, T. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by
Mims, A.
in:
PubMed
|
Google Scholar
|

1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Canfield, D. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Phillips, H. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Williams, K. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Shaffer, J. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Lozanski, A. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Doong, T. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Lozanski, G. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Mao, C. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Walker, C. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by
Blachly, J.
in:
PubMed
|
Google Scholar
|

1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Daniyan, A. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Alinari, L. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Baiocchi, R. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by
Yang, Y.
in:
PubMed
|
Google Scholar
|

1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Grieselhuber, N. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Campbell, M. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Baker, S. in: PubMed | Google Scholar
1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by
Blaser, B.
in:
PubMed
|
Google Scholar
|

1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by
Abdel-Wahab, O.
in:
PubMed
|
Google Scholar
|

1Division of Hematology, Department of Internal Medicine, The Ohio State University, Columbus, Ohio, USA.
2Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, USA.
3Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio, USA.
4University of Pennsylvania, Philadelphia, Pennsylvania, USA.
5Section of Hematology/Oncology, University of Chicago, Chicago, Illinois, USA.
6Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
7College of Veterinary Medicine, Michigan State University, East Lansing, Michigan, USA.
8Memorial Sloan Kettering Cancer Center, New York, New York, USA.
9Leukemia Research Program, The Ohio State University James Comprehensive Cancer Center, Columbus, Ohio, USA.
10Division of Cancer Biology, Cedars Sinai Medical Center, Los Angeles, California, USA.
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
Authorship note: BWB, OAW, and RL contributed equally to this work.
Find articles by Lapalombella, R. in: PubMed | Google Scholar
Authorship note: BWB, OAW, and RL contributed equally to this work.
Published March 20, 2025 - More info
J Clin Invest. 2025;135(10):e184021. https://doi.org/10.1172/JCI184021.
© 2025 Zhang et al. This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Received: June 20, 2024; Accepted: March 6, 2025
-
Abstract
Mutations and deletions in TP53 are associated with adverse outcomes in patients with myeloid malignancies, and there is an urgent need for the development of improved therapies for TP53-mutant leukemias. Here, we identified mutations in TET2 as the most common co-occurring mutation in patients with TP53-mutant acute myeloid leukemia (AML). In mice, combined hematopoietic-specific deletion of TET2 and TP53 resulted in enhanced self-renewal compared with deletion of either gene alone. Tp53/Tet2 double-KO mice developed serially transplantable AML. Both mice and patients with AML with combined TET2/TP53 alterations upregulated innate immune signaling in malignant granulocyte-monocyte progenitors, which had leukemia-initiating capacity. A20 governs the leukemic maintenance by triggering aberrant noncanonical NF-κB signaling. Mice with Tp53/Tet2 loss had expansion of monocytic myeloid-derived suppressor cells (MDSCs), which impaired T cell proliferation and activation. Moreover, mice and patients with AML with combined TP53/TET2 alterations displayed increased expression of the TIGIT ligand, CD155, on malignant cells. TIGIT-blocking antibodies augmented NK cell–mediated killing of Tp53/Tet2 double-mutant AML cells, reduced leukemic burden, and prolonged survival in Tp53/Tet2 double-KO mice. These findings describe a leukemia-promoting link between TET2 and TP53 mutations and highlight therapeutic strategies to overcome the immunosuppressive bone marrow environment in this adverse subtype of AML.
-
Introduction
Mutations in TP53 are independently associated with inferior survival in patients with myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) (1). TP53 mutations are present in 10%–15% of patients with AML, frequently co-occur with complex karyotype, and are more common in patients with secondary or therapy-related AML. Mutations in TP53 may occur years to decades before development of overt neoplasia and define a high-risk subtype of clonal hematopoiesis.
Several prior studies have evaluated genomic alterations that coexist with TP53-mutant myeloid neoplasms. Among these, loss of heterozygosity and deletion of the p53 locus on chromosome 17p are well described to co-occur with TP53 mutations (1–5). Beyond additional alterations to TP53, combined mutations in TP53 and TET2 are observed in MDS and are associated with high risk of AML transformation (6, 7). Moreover, TET2/TP53 mutation is an independent predictor of inferior survival after stem cell transplantation (8).
Given the profound adverse implications of TP53 mutations in myeloid neoplasms, there is an urgent need for targeted therapies to treat TP53-mutant leukemias and prevent the progression of these high-risk malignancies. Due to the impact of TP53 mutations on impairment of DNA damage response and induction of apoptosis there has been great hope that nontumor cell autonomous therapeutic strategies harnessing the immune system could be used to treat TP53-mutant leukemias. To this end, prior work has suggested that TP53-mutant MDS may be characterized by impairments in the function and abundance of a broad range of adaptive immune cells (9). In parallel, several studies have also documented increased expression of the immune checkpoints PD-1, PD-L1, and CTLA-4 on myeloid malignant cells in MDS and AML (9). Despite these observations, blockade of these specific immune checkpoints in MDS has failed to result in therapeutic benefit.
Herein, by studying two cohorts of patients with AML from different institutions, we found that 10% of TP53-mutant AML cases carry a concurrent TET2 mutation and are associated with extremely poor clinical outcomes. Our findings showed that mice with combined deletion of TET2 and TP53 developed aggressive acute leukemia that closely resembled human AML. Functional analyses of patients with TET2/TP53 comutant AML revealed activation of proinflammatory A20-mediated innate immune signaling in the malignant cells. We found clear evidence of T cell exhaustion, with upregulation of the TIGIT ligand, CD155, on malignant cells, both in mouse models and patients with AML harboring these comutations. Importantly, therapeutic blockade of the CD155-TIGIT axis with anti-TIGIT antibodies markedly promoted NK cell–mediated killing of TP53/TET2 comutant AML cells and extended survival in mice engrafted with Tp53/Tet2 double-KO AML cells. These findings provide evidence that TP53 deficiency and TET2 loss cooperate to drive the development of AML and that the immunosuppressive microenvironment seen in such cases may be amenable to therapeutic blockade of TIGIT signaling.
-
Results
Mutations in TET2 are frequent in TP53-mutant AML. We analyzed mutational data from 983 adult patients with TP53-mutant AML (216 from the Alliance for Clinical Trials in Oncology [referred to herein as Alliance]), the Beat AML program (10), and Rodriguez-Meira et al. (11) datasets, 99 from the University of Chicago, and 668 from American Association for Cancer Research (AACR) Project Genomics Evidence Neoplasia Information Exchange (GENIE) (12) to identify genetic alterations coexisting with TP53 mutations in AML. The spectrum of TP53 mutations in these cohorts is consistent with that in prior publications, with 50% consisting of a single nucleotide variant at hot spots R273, R248, H179, R175, and Y200 and the rest of the mutations being frameshift (28%) and nonsense (13%) mutations predicted to result in loss of function (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI184021DS1). Interestingly, the single most commonly comutated gene with TP53 was TET2, with 12% of patients with TP53-mutant AML having a coexisting TET2 mutation (Figure 1A and Supplemental Figure 1B). TET2 mutations occurred along the entire open-reading frame with a few notable hot spots at R550, R1261, and I1873, as previously reported (13) (Supplemental Figure 1C).
 Figure 1
Figure 1TET2 mutations are common in TP53-mutant AML and confer an inferior outcome. (A) Oncoprint of patients with TP53-mutant AML with indication of the top 6 most common co-occurring genetic events as well as cytogenetics and tumor mutational burden (TMB) among 216 patients with AML with somatic TP53 mutations. Data are from Alliance (our unpublished observations), Beat AML (10), and Rodriguez-Meira et al. (11). (B) Density estimation of variant allele frequency (VAF) of TP53 mutations across 668 TP53-mutant patients (subdivided by patients with 1 TP53 mutation, >1 TP53 mutation, or TP53 mutation plus deletion). Data are from AACR Project GENIE (12). (C) As in B but for TET2 mutations from 80 TP53-mutant patients (subdivided by patients with 1 TET2 mutation, >1 TET2 mutation, or TET2 mutation plus deletion). (D) The portion and VAF of TET2 and TP53 mutations in 26 patients with TP53/TET2 comutations. Data are from Alliance, Beat AML (10), and Rodriguez-Meira et al. (11). (E) Kaplan-Meier survival curve in 1,603 patients with AML based on TP53 and TET2 mutational status from Alliance. OS, overall survival. (F) As in E but for an independent cohort of 653 patients with AML from the University of Chicago. A log-rank test was used for survival statistics.
Prior studies have highlighted that the allelic state of TP53 is critical in determining prognosis in TP53-mutant MDS (13). While monoallelic TP53 mutations were mostly subclonal, biallelic TP53 mutations or TP53 mutations accompanied by deletions of the remaining allele were clonal (Figure 1B). Similarly, individual TET2 mutations in TP53-mutant AML were subclonal with a variant allele frequency (VAF) of 0.02, but biallelic TET2 mutations or mutations plus deletions in TET2 resulted in much higher VAFs and clonal dominance (Figure 1C). The spectrum of TP53 and TET2 mutations was similar along allelic states and had the same characteristic hot spot mutations (Supplemental Figure 1, A and C). Of these 26 cases with TP53 and TET2 comutations, 16 of them harbored 2 distinct TP53 mutations with VAFs around 0.5; 9 had mutations with VAFs close to 1; 1 of them had a single mutation with VAF around 0.5 or less (Figure 1D). The frequency of multihit TET2 mutations was much higher than that of single-hit TET2 mutations. Homozygous biallelic mutations (VAF > 0.9) and heterozygous biallelic mutations (2 mutations with VAF = 0.5) account for 80% of all TET2 mutation types. Biallelic mutation in TET2 was more frequent compared with monoallelic mutation in the multihit TP53 subgroup.
While the presence of TP53 mutations was associated with poor survival, patients harboring concurrent TP53 and TET2 mutations exhibited even shorter overall survival compared with TP53 or TET2 single-mutated patients in both the Alliance and University of Chicago datasets (P < 0.01) (Figure 1, E and F). Altogether, these data identify the frequent co-occurrence of TET2 mutations among TP53-mutant AML and suggest functional importance of loss of TET2 in this adverse prognostic group of patients with AML.
Loss of Tet2 and Tp53 expression results in lethal AML. To study the functional significance of loss of TET2 in the setting of TP53 loss-of-function mutations, we generated conditional mice with hematopoietic-specific deletion of both genes by crossing Vav-cre Tet2fl/fl and Tp53fl/fl mice. These animals mimicked the clonal combined biallelic mutations in TP53 and TET2 seen in patients with AML (Figure 1). Vav-cre Tet2fl/fl Tp53fl/fl double-KO mice were viable at birth and born at normal Mendelian ratios, but their median survival (21 weeks) was significantly shorter than that of littermate Vav-cre control (WT) or single-gene KO control mice (Figure 2A). In addition, the median survival was shorter than that for previously described Tet2–/–Flt3-ITD mice (37 weeks) (14). Complete differential blood counts revealed leukocytosis, anemia, and thrombocytopenia in 4-month-old Vav-cre Tet2fl/fl Tp53fl/fl mice (Figure 2, B and C). Necropsy of Vav-cre Tet2fl/fl Tp53fl/fl mice revealed splenomegaly (Supplemental Figure 1D) and infiltration of immature appearing cells in the liver, lung, lymph node, spleen, and bone marrow (Figure 2, D and E).
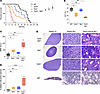 Figure 2
Figure 2Hematopoietic–cell specific Tp53- and Tet2-KO mice develop lethal leukemia. (A) Kaplan-Meier survival curves of Tp53–/–Tet2–/–, Tet2–/–, Tp53–/–, and WT mice. n = 32 WT mice, n = 23 Tp53–/– mice, n = 19 Tet2–/– mice, and n = 21 Tp53–/–Tet2–/– mice. **P < 0.01; ***P < 0.001. (B and C) Box-and-whisker plots of (B) hematocrit (HCT) and (C) white blood cell (WBC) counts of 4-month-old mice with different genotypes. (D) H&E staining of spleen and bone marrow in moribund mice of various genotypes. Scale bar: 100 μm; original magnification, ×2 (spleen, left); ×60 (spleen, right, and bone marrow). Data are representative of n = 6–7 mice/genotype. (E) Box-and-whisker plots of percentage of bone marrow blasts, based on D at the age of 4 months. For box-and-whisker plots in B, C, and E, boxes represent median, first, and third quartiles, with whiskers extending to 1.5× interquartile range. n = 6–7 mice/genotype. ANOVA with Dunnett’s test was used for significance. *P < 0.05; **P < 0.01; ***P < 0.001. Tp53–/–Tet2–/–, Vav-cre Tet2fl/fl Tp53fl/fl; Tet2–/–, Vav-cre Tet2fl/fl; Tp53–/–, Vav-cre Tp53fl/fl; WT, Vav-cre.
Vav-cre Tet2fl/flTp53fl/fl mice had myeloid lineage profiles of leukemia, with dominant cell populations expressing CD11b and Gr-1, but not a pan–B cell (B220) or a T cell marker (CD3) (Figure 3A). Splenocytes of Vav-cre Tet2fl/fl Tp53fl/fl mice that developed AML had substantially increased frequencies of Cd11b+Gr-1– and Cd11b+Gr-1+ myeloid cells as well as increased CD11b+cKIT+ cells (Figure 3, A and B). Moreover, an abnormally high frequency of cKIT+ cells could be detected in peripheral blood of Vav-cre Tet2fl/fl Tp53fl/fl mice by 4 months of age (Figure 3C). IHC staining of Vav-cre Tet2fl/fl Tp53fl/fl moribund mice that developed AML showed results that were consistent with flow cytometry analysis, revealing myeloid cell expansion and nearly absent lymphoid cells in spleen (Figure 3D). Consistent with previous reports, expressions of CD3 and Sca-1 were increased in Vav-cre Tp53fl/fl moribund mice developing T acute lymphoblastic leukemia (Figure 3D) (15).
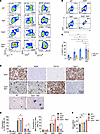 Figure 3
Figure 3Tp53/Tet2 double-KO mice develop AML characterized by expansion of granulocyte macrophage progenitors. (A) Representative flow cytometry analysis of lineages of CD45dimSSClo cells in spleens from Tp53–/–Tet2–/–, Tet2–/–, Tp53–/–, and WT mice at the time of sacrifice (4 months of age). (B) Representative flow cytometry analysis of CD11b+cKIT+ cells in peripheral blood from moribund 4-month-old Tp53–/–Tet2–/– mice and age-matched controls. (C) Frequency of CD11b+ and cKIT+ cells among CD45.2+ cells in peripheral blood of moribund 4-month-old Tp53–/–Tet2–/– mice and age-matched controls. n = 3–6 mice/group. *P < 0.05. Mean ± SEM. ANOVA with Dunnett’s test was used for significance. (D) Top: Immunohistochemical staining of spleens of representative moribund mice for the proteins indicated. Scale bar: 100 μm. Bottom: Wright-Giemsa stain of peripheral blood of Tp53–/–Tet2–/– and littermate WT mice. Data are representative of n = 6 mice/group. Scale bar: 10 μm. (E) Left: Frequencies of long-term hematopoietic stem cells (LT-HSC), multipotent progenitors (MPP), and short-term HSCs (ST-HSC) among bone marrow lineage-negative Sca1+cKIT+ (LSK) cells of 16-week-old mice with the indicated genotypes. Right: Frequencies of common myeloid progenitors (CMP), granulocyte macrophage progenitors (GMPs), and megakaryocyte-erythroid progenitors (MEPs) among bone marrow Lin–Sca-1–cKIT+ cells of 16-week-old mice with the indicated genotypes. n = 3 mice/group; Mean ± SD. ANOVA with Dunnett’s test. (F) Percentage of bone marrow EdU+ GMPs in 16-week-old mice with the indicated genotypes. n = 5 mice/group. *P<0.05. ANOVA with Dunnett’s test was used for significance. Tp53–/–Tet2–/–, Vav-cre Tet2fl/fl Tp53fl/fl; Tet2–/–, Vav-cre Tet2fl/fl; Tp53–/–, Vav-cre Tp53fl/fl; WT, Vav-cre.
Given the known role of Tet2 in regulating frequency and self-renewal of hematopoietic stem and progenitor cells (HSPCs) (14), we analyzed bone marrow HSPCs in Vav-cre Tet2fl/fl Tp53fl/fl mice and controls at 4 months of age. The frequency of multipotent progenitor cells was significantly increased in Vav-cre Tet2fl/fl Tp53fl/fl mice relative to that in other groups, while short-term hematopoietic stem cells (HSCs) were slightly reduced (Figure 3E). We observed an expansion of granulocyte-macrophage progenitors (GMPs), with a marked reduction of megakaryocyte-erythroid progenitors (MEPs) in Vav-cre Tet2fl/fl Tp53fl/fl mice compared with that in other groups (Figure 3E). Based on in vivo EdU labeling, GMPs in Vav-cre Tet2fl/fl Tp53fl/fl mice with AML were more proliferative than those in controls (Figure 3F). Altogether, we found that concurrent loss of Tet2 and Tp53 cooperatively results in development of lethal AML and alters HSPC frequencies.
Combined deletion of Tp53 and Tet2 enhances HSPC self-renewal. Deletion of Tet2 has been repeatedly shown to enhance HSPC self-renewal in vitro and in vivo (16, 17). We therefore next examined the impact of combined Tet2 and Tp53 deletion on HSPC self-renewal. Bone marrow cells from Vav-cre Tet2fl/fl Tp53fl/fl mice sustained up to 5 rounds of plating, at which time they yielded larger numbers of cells in culture relative to controls (Figure 4A). To evaluate self-renewal in vivo, we conducted competitive transplantation by transplanting 1 × 106 bone marrow cells from CD45.2+ Vav-cre Tet2fl/fl Tp53fl/fl, Vav-cre Tet2fl/fl, Vav-cre Tp53fl/fl, or Vav-cre control mice together with congenic CD45.1+ competitor cells at 1:1 ratio into lethally irradiated CD45.1+ recipient mice (Figure 4B). At these cell doses, recipients of Vav-cre Tet2fl/fl Tp53fl/fl cells developed lethal myeloid neoplasms and died by 33 days after transplantation (Figure 4B). Robust engraftment was observed, with 30%–60% of CD45.2+ donor cells readily detectable in peripheral blood of recipients of Vav-cre Tet2fl/fl Tp53fl/fl bone marrow cells as early as 3 weeks after engraftment (Figure 4C). Unlike mice engrafted with Vav-cre Tet2fl/fl Tp53fl/fl cells, mice receiving Vav-cre Tet2fl/fl or Vav-cre Tp53fl/fl donor cells had longer survival times and died from a chronic myelomonocytic leukemia–like disease (for Vav-cre Tet2fl/fl mice) or T cell malignancies (for Vav-cre Tp53fl/fl mice) (Figure 4D).
 Figure 4
Figure 4Increased hematopoietic progenitor self-renewal in Tp53/Tet2 double-KO mice. (A) Number of colonies from plating of 10,000 cells from the bone marrow of 16-week-old Vav-cre Tp53fl/flTet2fl/fl mice and controls in methylcellulose. Mean ± SD of 3 technical replicates. (B) Kaplan-Meier curves of recipient CD45.1+ mice following competitive transplantation of bone marrow cells (1 × 106 cells) from leukemic CD45.2+ primary transgenic mice with the indicated genotypes into lethally irradiated recipient mice with CD45.1+ supporting bone marrow cells (1 × 106 cells). n = 10 WT mice, n = 10 Tp53–/– mice, n = 12 Tet2–/– mice, and n = 10 Tp53–/–Tet2–/– mice. (C) Box-and-whisker plots of CD45.2+ cells in peripheral blood of mice from B. Boxes represent median, first, and third quartiles, with whiskers extending to 1.5× interquartile range. n = 10 mice/genotype. (D) Disease incidence in moribund recipient mice following competitive transplantation of bone marrow cells from primary transgenic mice with the indicated genotypes. T-ALL, T acute lymphoblastic leukemia; CMML, chronic myelomonocytic leukemia; AML, acute myeloid leukemia. Tp53–/–Tet2–/–, Vav-cre Tet2fl/fl Tp53fl/fl; Tet2–/–, Vav-cre Tet2fl/fl; Tp53–/–, Vav-cre Tp53fl/fl; WT, Vav-cre. A log-rank test was used for survival statistics; otherwise, ANOVA with Dunnett’s test was used for P values. *P < 0.05; **P < 0.01; ***P < 0.001.
TLR2/A20/noncanonical NF-κB pathway–mediated proinflammatory signaling promotes myeloid leukemia development in TET2/TP53-mutant progenitors. To understand the molecular mechanisms underlying AML transformation upon combined mutation in TET2 and TP53, we performed bulk RNA-Seq of Lin–cKIT+ myeloid progenitors (LK) and Lin–cKIT+Sca-1+ (LSK) cells from diseased Vav-cre Tet2fl/fl Tp53fl/fl mice developing AML and age-matched Vav-cre Tp53fl/fl, Vav-cre Tet2fl/fl, and WT mice. The myeloid transcriptional factors RUNX1, SPI1, GFI1, PPARG, LBR, and CITED2 were upregulated in expression in Vav-cre Tet2fl/fl Tp53fl/fl LSK cells compared with other groups of cells (Figure 5A, Supplemental Figure 2A, and Supplemental Table 1), and gene set enrichment analysis (GSEA) revealed an enrichment in gene signatures in LSK cells related to myeloid differentiation and AML transformation (such as immortalized HOXA9, MEIS1_up, GATA2 targets-up, and myeloid cell development up) (Supplemental Figure 2B). Additionally, gene signatures associated with Myc and inflammatory responses were enriched in Vav-cre Tet2fl/fl Tp53fl/fl AML versus WT LSK cells (Figure 5B). In contrast, T lymphocyte commitment gene sets were enriched in Vav-cre Tp53fl/fl LSK progenitors, consistent with findings that Tp53 loss alone in Vav-cre Tp53fl/fl mice primarily drove T acute lymphoblastic leukemia (Supplemental Figure 2, C and D).
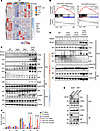 Figure 5
Figure 5Myeloid predisposition and enhanced innate immune signaling in Tp53–/–Tet2–/– precursor cells. (A) Heatmap of top differentially expressed genes (FDR < 0.05) within the myeloid differentiation pathway of LSK cells from 4-month-old mice with different genotypes. Key myeloid transcriptional factors upregulated in Tp53/Tet2 double-KO LSK cells relative to other groups are highlighted. (B) GSEA plots of inflammatory responses and the Myc pathway in LSK cells from Tp53–/–Tet2–/– mice with AML relative to control mice. (C) Western blot of TLR2, A20, noncanonical NF-κB pathway components (NIK, p52/p100, and phosphorylated p100 [p-p100] in whole-cell lysate [WCL] and RelB in nuclear extract [NE]) and canonical NF-κB pathway members (p65, phosphorylated-IKKα [p-IKKα], and IKKα) in cKit+ bone marrow cells from age-matched mice with indicated genotypes (3 mice per group). β-Actin and Lamin B1 served as housekeeping controls in WCL and NE, respectively. (D) Western blot of TLR2, A20, NIK, and p52/p100 in WCL and RelB in NE from patients with combined TET2 and TP53 mutations, either mutation alone, or neither TET2 or TP53 mutations (WT). β-Actin and Lamin B1 served as housekeeping controls in WCL and NE, respectively. Analysis of fold change normalized to the control lane is shown below immunoblot where indicated. (E) Western blot of A20, phosphorylated IkBα (p-IkBα), and total IkBα in WCL and RelB and RelA in NE in cKit+ bone marrow cells of Tp53–/–Tet2–/– mice treated with control (sgNeg) or 1 of 2 A20-targeting sgRNAs. β-Actin and Lamin B1 served as housekeeping controls in WCL and NE, respectively. (F) Mean number of methylcellulose colonies in cells from E and WT bone marrow cells treated with control or 1 of 2 A20-targeting sgRNAs. Mean ± SD shown. **P < 0.01. Results are representative of 2–3 independent experiments. Tp53–/–Tet2–/–, Vav-cre Tet2fl/fl Tp53fl/fl; Tet2–/–, Vav-cre Tet2fl/fl; Tp53–/–, Vav-cre Tp53fl/fl; WT, Vav-cre.
Beyond genes involved in myeloid cell commitment and oncogenes, we observed increased expression of Tnfaip3 and Tlr2 in Vav-cre Tet2fl/fl LK cells, compared with WT LK cells, which was further increased in Vav-cre Tet2fl/fl Tp53fl/fl LK cells (Supplemental Figure 2E). Tnfaip3 encodes A20 deubiquitinase, and Tlr2 encodes TLR2. A20 can mediate HSC transformation by activating noncanonical NF-κB signaling (17). Inhibition of A20 expression can prevent TLR-TRAF6–primed TET2-mutant MDS from progressing to leukemia (18). Thus, we evaluated A20 and associated canonical/noncanonical NF-κB pathway component protein levels. Our data showed increased levels of TLR2, A20, and noncanonical NF-κB pathway components, including NIK, p100, p52, phosphorylated p100, and elevated nuclear-localized RelB, in Vav-cre Tet2fl/fl Tp53fl/fl mice in comparison to Vav-cre Tet2fl/fl, Vav-cre Tp53fl/fl or WT mice (Figure 5C). In contrast, canonical NF-κB pathway members p65, phosphorylated IKKα, and IKKα were not upregulated in Vav-cre Tet2fl/fl Tp53fl/fl mice (Figure 5C). Similarly, TLR2, A20, NIK and the ratio of p52/p100 were consistently elevated in bone marrow or blood cells from patients with AML harboring comutations in TET2 and TP53 (Figure 5D and Supplemental Table 2). In addition, we observed more nuclear-localized RelB in cells from patients with TP53/TET2 comutations compared with those with single mutations (Figure 5D).
Previous studies have suggested that TET2-deficient HSPCs express A20 and exhibit noncanonical NF-κB activation. Additionally, LPS can enhance the expansion of Tet2-KO HSCs by activating NF-κB signaling (18, 19). Furthermore, LPS promotes malignant transformation in Tet2-mutant mice by accelerating the production of MHC IIhi monocytes (20). Infection-derived LPS drives preleukemic myeloproliferation in these mice, a process that can be reversed by antibiotic treatment (19). To assess TLR function in leukemia cells, we measured in vitro responses of mouse cells to the TLR2 agonist, PAM3CSK4. Upon PAM3CSK4 stimulation, the levels of A20, NIK, and phosphorylated p100 and the ratio of p52/p100 were elevated in cells derived from WT and single-mutant mice, as expected (Supplemental Figure 2F). However, the extent of upregulation of these proteins was substantially higher in Vav-cre Tet2fl/fl Tp53fl/fl cells compared with WT or single-mutant cells. These findings indicate that the noncanonical NF-kB signaling pathway was hyperactivated in response to TLR2 stimulation in Vav-cre Tet2fl/fl Tp53fl/fl murine hematopoietic cells (Supplemental Figure 2F). In contrast, canonical NF-κB pathway components, including p50, IkBα, and phosphorylated IkBα, did not exhibit differential changes in response to TLR2-induced activation in Vav-cre Tet2fl/fl Tp53fl/fl cells (Supplemental Figure 2F).
The data above suggest that combined loss of TET2 and TP53 enhances sensitivity to innate immune signaling, revealing cooperative upregulation of myeloid transcription factor expression and enhancement of proinflammatory signaling. We next sought to determine the functional requirement for A20 in the maintenance of Tp53/Tet2 double-mutant leukemia. We genetically depleted A20 in mouse cKit+ bone marrow cells from Vav-cre Tet2fl/fl Tp53fl/fl mice (Figure 5E). A20 deletion was confirmed by Western blot. Deletion of A20 decreased the level of nuclear RelB while increasing nuclear localization of p65, and promoted phosphorylation of IkBα (Figure 5E). Additionally, A20 loss strikingly reduced the serial replating capacity of Tp53/Tet2 double-KO murine AML cells in vitro (Figure 5F).
Tumor-intrinsic transcriptional features are associated with distinct leukemia phenotypes. Given the heterogeneity of malignant cells in leukemia, we next performed single-cell RNA-Seq (scRNA-Seq) of bone marrow cells from Vav-cre Tet2fl/fl Tp53fl/fl mice as well as WT and Vav-cre Tp53fl/fl mice. Uniform manifold approximation and projection (UMAP) dimension reduction of all samples yielded 16 distinct partition clusters (Figure 6A). Cells in clusters 3 and 6, predominantly contributed by Vav-cre Tet2fl/fl Tp53fl/fl AML cells, showed enrichment of myeloid leukemia marker genes as well as myeloid-associated genes (Figure 6, B and C) (21). To further evaluate intratumoral heterogeneity in TP53/TET2 comutant AML, we further fragmented AML clusters 3 and 6 and identified 5 distinct clusters of AML cells, including OXPHOShi, IFNhi, hyperproliferative, neutrophil-like, and erythroid-like AML populations (Figure 6, D and E) (22).The IFNhi AML population displayed enrichment in myeloid suppressor cell markers, macrophage myeloid differentiation markers (Ccl6), the eosinophil IgE receptor (Fcer1g), and Cd52, which is highly expressed on leukemia progenitor cells (Figure 6E). Endosome-associated factor, Ifitm3, showed aberrantly high expression in the IFNhi cells (Figure 6E). Genes associated with secondary granules were upregulated in neutrophil-like AML (23). The hyperproliferative AML subcluster displayed features of G1/S phase transition with histone gene expression and mitotic drivers. OXPHOShi AML cells express a variety of mitochondrial respiratory chain–related genes (24). Erythroid-like AML (cluster 6) is characterized by the expression of hemoglobin genes alongside myeloid markers such as Elane, Mpo, and Cd34. However, this erythroid-like cluster differs from acute erythroid/megakaryocytic leukemia, which often originates from the common MEP. In Vav-cre Tet2fl/fl Tp53fl/fl mice, MEPs were notably reduced, correlating with the anemia and thrombocytopenia observed in these mice (Figure 2B). Clusters 3 and 6, as well as the associated 5 subclusters were negative for lineage genes but coexpressed HSC/multipotent progenitor cell markers Cd48, Cd150, Kit, and Sca1 (Supplemental Figure 3, A and B).
 Figure 6
Figure 6Tp53/Tet2 comutant AML displays unique transcriptional signatures. (A) UMAP plots of single-cell transcriptomes of bone marrow mononuclear cells from Vav-cre control mice (WT), Vav-cre Tp53fl/fl mice, or Vav-cre Tet2fl/fl Tp53fl/fl mice. Cell density (2D kernel density estimate mapped to color scale) plots are shown underneath. n = 2 mice/group; 3 weeks after engraftment into CD45.1+ mice. (B) UMAP projection of the expression of selected myeloid marker genes. Expression is displayed by color scale as log10-transformed expression (size factor normalized unique molecular identifier counts). (C) Heatmap with hierarchical clustering showing top differentially expressed genes in clusters from A across the different groups of mice. The normalized proportion allocations of cells of various phenotypes in each cluster are shown as stacked bar plots underneath the heatmap. (D) Annotated subpopulations (Leiden clusters) of AML blast clusters (partition clusters 3 and 6) (49) showing identified AML subtypes. (E) Heatmap showing top differentially expressed genes identified in each AML subtype from D. Tp53–/–Tet2–/–, Vav-cre Tet2fl/fl Tp53fl/fl;Tp53–/–, Vav-cre Tp53fl/fl; WT, Vav-cre.
To evaluate the similarity between mouse and human AML, we performed cellular indexing of transcriptome and epitope sequencing (CITE-Seq) with 12 patients with TP53/TET2-mutant AML, 6 patients with TP53-mutant AML, 6 patients with TET2-mutant AML, and 6 patients with WT AML and projected mouse scRNA-Seq data onto human AML CITE-Seq data (Supplemental Figure 3, C and D). This comparative analysis revealed that Vav-cre Tet2fl/fl Tp53fl/fl murine AML cells significantly align transcriptionally with TP53/TET2 comutant human AML cells compared with single-mutant or WT control cells (binomial test, P < 2.2 × 10–16, predicted probability = 0.25, observed probability = 0.78, 95% confidential interval = 0.76) (Supplemental Figure 3, E and F). This suggests that distinct clusters 3 and 6 of Vav-cre Tet2fl/fl Tp53fl/fl murine AML closely mirror the cellular heterogeneity and transcriptional signatures observed specifically in the corresponding TP53/TET2 comutant human AML.
Cluster 1, which was enriched in wild-type, single-KO, and double-KO cells, showed high expression of B cell lineage marker genes, Cd19, Cd24a, Pax5, and Cd79a, and genes with functions in class-switch recombination and somatic hypermutation (Figure 6C and Supplemental Figure 4A). In contrast, T cell markers were expressed in cluster 2, which is specific for Vav-cre Tp53fl/fl cells (Supplemental Figure 4B).
Next, we evaluated molecular mechanisms that may drive different leukemia fates upon combined TET2 and TP53 deletion. Vav-cre Tet2fl/fl Tp53fl/fl AML bone marrow cells formed a distinct cluster compared with Vav-cre Tp53fl/fl and WT mouse–derived cells (Louvain cluster 19; Supplemental Figure 5, A and B). The cells in cluster 19 were predominantly from clusters 3 and 6 (Figure 6A). The expression levels of Klf4 and Itga4 were progressively elevated during transition from early precursors to AML, peaking in cluster 19 (Supplemental Figure 5C).
GMPs in Tp53 and Tet2 double-KO AML exhibit distinct transcriptional signatures and acquire leukemia-initiating capacity. Given the expansion of GMPs in Vav-cre Tet2fl/fl Tp53fl/fl mice, we also sought to evaluate gene expression specifically in Vav-cre Tet2fl/fl Tp53fl/fl GMPs relative to controls. We therefore sorted GMPs from Vav-cre Tet2fl/fl Tp53fl/fl mice or control mice and performed RNA-Seq. Gene expression of GMPs from Vav-cre Tet2fl/fl Tp53fl/fl mice was profoundly altered in comparison to that in single-mutant controls. Compared with Vav-cre Tet2fl/fl GMPs, MHC class II genes, H2-Aa and H2-Eb1, and MHC II trafficking adaptor gene, Cd74, were significantly downregulated in Vav-cre Tet2fl/fl Tp53fl/fl GMPs (Figure 7A and Supplemental Table 3). In sharp contrast, stem cell gene, Cd34; lineage gene, Gata2; and leukemia stem cell regulator, Ikzf2; as well as Tnfaip3, Pvr (Cd155), and Nectin2 (Cd112), were upregulated in Vav-cre Tet2fl/fl Tp53fl/fl GMPs (Figure 7A). In comparison to Vav-cre Tp53fl/fl GMPs, tumor suppressor/p21 stabilizer, Rbms2, was downregulated, while transcripts of stem cell genes, Cd34 and Cd44; leukemia enhancer genes, Runx1, Ikzf2, and Gata2; inflammasome gene, Nlrp1b; and Tnfaip3, Pvr (Cd155), and Traf2, were significantly upregulated in GMPs from Vav-cre Tet2fl/fl Tp53fl/fl mice (Figure 7B and Supplemental Table 4). Moreover, gene expression signatures of noncanonical NF-κB signaling, MYC targets, and genes enriched in normal HSCs versus GMPs were positively enriched in Vav-cre Tet2fl/fl Tp53fl/fl GMPs compared with those from single-KO GMPs (Figure 7, C and D, and Supplemental Tables 5 and 6). Conversely, genes associated with NK cell–mediated immunity were negatively enriched in Vav-cre Tet2fl/fl Tp53fl/fl GMPs. These observations are consistent with our findings in LK cells (Figure 5B).
 Figure 7
Figure 7Mutant Tp53 and Tet2 cooperatively transform murine GMP progenitors. Volcano plots depicting top differentially expressed genes in GMPs (Lin–cKIT+Sca1– CD16/32+ CD34+) from (A) Tp53–/–Tet2–/– versus Tet2–/– or (B) Tp53–/–Tet2–/– versus Tp53–/– mice. (C) GSEA plots of pathways enriched in GMPs from Tp53–/–Tet2–/– mice with AML relative to Tet2–/– control mice. (D) As in C but for GMPs from Tp53–/–Tet2–/– mice with AML relative to Tp53–/– control mice. (E) Heatmap of top differentially expressed genes (adjusted P < 0.05) within the STEMNESS_UP pathway of GMP cells from 4-month-old mice with different genotypes. Key genes upregulated in Tp53–/–Tet2–/– GMP cells are highlighted. (F) CD45.2+ (mutant) to CD45.1+ (WT) cell ratios and (G) CD11b+Gr1+ percentage in CD45.2+ cells in the peripheral blood of recipient mice engrafted with CD45.2+ cell types (either GMPs or whole bone marrow [BM]) from the animals with the indicated genotypes. For box-and-whisker plots, boxes represent median, first, and third quartiles, with whiskers extending to 1.5× interquartile range. P values are shown (ANOVA with Dunnett’s test). ***P < 0.001; **** P < 0.0001. (H) Kaplan-Meier curves of lethally irradiated CD45.1+ mice receiving whole BM or GMP cells from mice with different genotypes. Log-rank test was used for survival statistics. *P < 0.05. Tp53–/–Tet2–/–, Vav-cre Tet2fl/fl Tp53fl/fl; Tet2–/–, Vav-cre Tet2fl/fl; Tp53–/–, Vav-cre Tp53fl/fl; WT, Vav-cre.
Tp53 loss and Tet2 depletion cooperatively upregulated transcriptional factors regulating stemness (25), including Mdfic, Zfp54, Zfx, and Rnf4; chromatin-remodeling helicases of the SNF2/SWI2 family, including Chd1 and Smarcad1; and other well-known regulators of leukemogenesis, such as Lsm1, Xpo1, Kras, and Gnb1, in GMP progenitors (Figure 7E and Supplemental Table 7). Furthermore, Myc was elevated in GMPs in Vav-cre Tet2fl/fl Tp53fl/fl mice (Supplemental Figure 6A).
Compared with WT GMPs, Vav-cre Tet2fl/fl Tp53fl/fl GMPs exhibited a greater number of differentially expressed genes than the single-KO controls (Supplemental Figure 6B). Of these differentially expressed genes, 1,054 were upregulated and 1,616 were downregulated, with these changes being unique to the Vav-cre Tet2fl/fl Tp53fl/fl GMPs. This suggests a cooperative effect between Tp53 depletion and Tet2 loss.
The enrichment of stemness gene signature in GMPs from Vav-cre Tet2fl/fl Tp53fl/fl mice suggested that these cells may acquire self-renewal and leukemia-initiating cell features. To test this hypothesis, we sorted CD45.2+ GMPs from WT, Vav-cre Tet2fl/fl, Vav-cre Tp53fl/fl, and Vav-cre Tet2fl/fl Tp53fl/fl mice and engrafted them into lethally irradiated recipient mice with CD45.1+ supporting bone marrow mononuclear cells. We also included a positive control cohort of CD45.1+ mice engrafted with whole bone marrow mononuclear cells from Vav-cre Tet2fl/fl Tp53fl/fl mice. Four weeks after transplant, higher percentages of myeloid CD45.2+ cells were found in mice engrafted with Vav-cre Tet2fl/fl Tp53fl/fl GMPs or whole bone marrow compared with other groups (Figure 7, F and G). Moreover, transplantation of GMPs from Vav-cre Tet2fl/fl Tp53fl/fl mice resulted in death of recipient animals at a rate comparable to that of recipient mice engrafted with unfractionated bone marrow mononuclear cells (Figure 7H). These data indicated that Vav-cre Tet2fl/fl Tp53fl/fl GMP cells have leukemia-initiating capacity.
TP53 and TET2 double-mutant AML is characterized by T/NK cell exhaustion and monocytic MDSC-like cell expansion. One recent study proposed that mutant TP53 creates an immunosuppressive bone marrow environment in patients with MDS (26). We found that concomitant loss of Tp53 and Tet2 promotes expansion of CD11b+Ly6C+Ly6G– monocytic myeloid-derived suppressor cell–like (MDSC-like) cells but not CD11b+Ly6C–Ly6G+ granulocytic MDSC-like cells (Figure 8, A and B). When isolated and cocultured with T cells isolated from spleens of WT mice, monocytic MDSCs suppressed T cell proliferation in vitro (Figure 8, B and C). In addition, IFN-γ and TNF-α production was reduced in T cells cocultured with MDSCs derived from Vav-cre Tet2fl/fl Tp53fl/fl AML. In contrast, MDSCs from WT or Vav-cre Tet2fl/fl or Vav-cre Tp53fl/fl mice had less of an impact on T cell activation (Figure 8C).
 Figure 8
Figure 8Monocytic MDSCs emerge in Tp53–/–Tet2–/– leukemic environment to cause T cell dysfunction. (A) Representative FACS plots of the frequency of monocytic (CD11b+Ly6G–Ly6C+) and granulocytic MDSCs (CD11b+Ly6G+Ly6C–) in splenocytes from Vav-cre Tet2fl/flTp53fl/fl (Tp53–/–Tet2–/–), Vav-cre Tet2fl/fl (Tet2–/–), Vav-cre Tp53fl/fl (Tp53–/–), and WT mice (n = 6 mice/group). Suppression of T cell proliferation by isolated MDSCs: T cells from WT mouse spleens were isolated and stimulated with anti-CD3/CD28 Dynabeads. Subsequently, T cells were cocultured with MDSCs isolated from Tp53–/–Tet2–/–, Tet2–/–, Tp53–/–, and WT mouse spleens for 72 hours. Cell division was measured with CFSE proliferation assays. n = 6 mice/group. (B) Quantification of the percentage of monocytic and granulocytic MDSCs in CD45.2+ splenocytes across genotypes, as in A. (C) Quantification of the percentage of CFSE dilution in T cell proliferation and percentage of cytokine-expressing T cells upon coculture with MDSCs isolated from Tp53–/–Tet2–/–, Tet2–/–, Tp53–/–, and WT mice. P values are shown (ANOVA with Dunnett’s test). *P < 0.05; **P < 0.01; ****P < 0.0001. (D) Representative flow plot showing the division of CD4+ or CD8+ T cells with CFSE staining in response to MDSC coculture in the presence or absence of blocking antibodies or arginase inhibitor (Nor-NOHA). cKit+CD11b+Ly6C+Ly6G– monocytic MDSCs from Tet2–/–Tp53–/– mice were sorted and cocultured with anti-CD3/CD28–activated congenic CD8+ or CD4+ T cells at 1:8 ratio in the presence of IL-2 for 72 hours. In the selected conditions, coculture was performed in the presence of blocking antibodies or Nor-NOHA (N-Hydroxy-nor-L-arginine). (E) Percentage of CFSE dilution in T cell proliferation in T cell–MDSC coculture under different treatment conditions. For box-and-whisker plots, boxes represent median, first, and third quartiles, with whiskers extending to 1.5 × interquartile range. n = 6 mice/group. P values are shown (ANOVA with Dunnett’s test). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Tp53–/–Tet2–/–, Vav-cre Tet2fl/fl Tp53fl/fl; Tet2–/–, Vav-cre Tet2fl/fl; Tp53–/–, Vav-cre Tp53fl/fl; WT, Vav-cre.
To understand whether the suppressive roles of monocytic MDSCs are mediated via soluble factors or direct receptor-ligand engagement, the MDSC and T cell coculture experiments above were repeated in the presence or absence of neutralizing antibodies against IL-10, TGF-β, and PD-L1 or Nor-NOHA (N-Hydroxy-nor-L-arginine) to inhibit arginase in MDSCs. CD4+ or CD8+ T cells were stained with CFSE, and the impact of MDSC coculture in the presence or absence of neutralizing antibodies or Nor-NOHA was evaluated. This assay clearly revealed that monocytic MDSCs suppressed the proliferation of CD4+ or CD8+ T cells (Figure 8, D and E). Importantly, however, the monocytic MDSC-mediated inhibition of CD8+ T cell proliferation and activation (indicated by elevated IFN-γ and TNF-α production) was rescued fully by Nor-NOHA (and only partially by anti–IL-10 or anti–TGF-β antibodies) (Figure 8E and Supplemental Figure 7A). Monocytic MDSC-mediated inhibition of CD4+ T cell proliferation and activation was rescued by anti–PD-L1 antibodies (Figure 8E and Supplemental Figure 7A). These data suggest that monocytic MDSC-mediated CD8+ T cell dysfunction is mediated via a soluble factor (likely via L-arginine deprivation), while the suppressive effect on CD4+ T cells is mediated via direct PD1–PD-L1 engagement. In agreement with in vitro assays, there were more TIGIT+LAG3+CTLA4+ dysfunctional T cells but fewer TIGIT–LAG3–CTLA4– CD3+ normal T cells in TP53/TET2 comutant patient samples (Supplemental Figure 7, B and C). To assess the presence of monocytic MDSCs in humans, we performed spectral flow cytometry utilizing markers, such as CD66b, CD16, CD33, CD11b, CD14, and CD15. Our findings indicate that, while granulocytic MDSCs were elevated in patients harboring TP53 mutations, monocytic MDSCs were significantly increased in individuals with concurrent TP53 and TET2 mutations (Supplemental Figure 8, A–C). In agreement with spectral flow analysis, CITE-Seq analysis uncovered that, at the single-cell level, the frequency of monocytic MDSCs (CD33+CD11b+HLA-DRlo/–CD14+CD15– cells) was drastically elevated in the microenvironment of AML with TP53 and TET2 comutations (Supplemental Figure 8, D–H). This suggests that an increased abundance of monocytic MDSCs is a characteristic feature in both murine and human AML harboring TP53 and TET2 comutations.
To further evaluate the immune cell composition of Vav-cre Tet2fl/fl Tp53fl/fl mice, T, B, and NK cell identities were further classified using ScType (27). Cells consistent with naive B cells, immature B cells, and naive CD4+ T cells were present in AML (Supplemental Figure 9, A and B). In contrast to WT mice, effector CD8+ T, NK, and CD8+ NKT cells were depleted in AML mice (Supplemental Figure 9, C–E). Among T cell populations, a subset of Cd3e+Tigit+Pd1+Lag3+ T cells, indicative of exhaustion, was identified in samples from Vav-cre Tet2fl/fl Tp53fl/fl AML mice (Supplemental Figure 9, F and G, and Supplemental Figure 10, A and B). These cells exhibited upregulation of the transcription factor, Tox, further supporting their exhausted phenotype in AML mice with Tp53 and Tet2 double-KO (Supplemental Figure 10, A and B) (28–30). Consistent with mouse scRNA-Seq, patient CITE-Seq analysis revealed that CD8+ T cells in TP53 and TET2 comutant patients exhibited increased expression of TOX, CD244, TIGIT, TIM3, and LAG3, which are key features of T cell exhaustion (Supplemental Figure 10, C and D). Bulk RNA-Seq of LK cells revealed genes encoding immune checkpoint receptors Cd47, CD112, and Cd155, among the top differentially expressed genes in Vav-cre Tet2fl/fl Tp53fl/fl AML. Flow cytometry confirmed that Cd155 and Cd47 were strongly expressed in cKIT+ leukemia progenitors from Vav-cre Tet2fl/fl Tp53fl/fl AML mice compared with cells from Vav-cre Tet2fl/fl, Vav-cre Tp53fl/fl, or WT mice (Figure 9, A–C, and Supplemental Figure 11A).
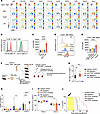 Figure 9
Figure 9Immunosuppressive microenvironment of TP53/TET2 double-mutant AML that is partially alleviated by TIGIT inhibition. (A) Representative t-distributed stochastic neighbor embedding (t-SNE) plots of multicolor flow cytometric analysis of selected immune checkpoint molecule levels on murine splenocytes from mice with the indicated genotypes. (B) Histograms of CD155 and CD112 expression by flow cytometry on LK cells of the bone marrow of Tp53–/–Tet2–/– or Cre+ control mice (WT). (C) Quantification of CD155 on murine cKIT+ progenitors of different genotypes by flow cytometry. Values (mean fluorescence intensity [MFI]) are shown as mean ± SEM; n = 3 mice/group; P values are shown (ANOVA with Dunnett’s test). ****P < 0.0001. (D) Representative flow plots and (E) quantification of CD155 expression on CD45dimSSCloCD33+ blasts from patients with AML harboring different mutations by spectral flow. Values (mean fluorescence intensity MFI) are shown as mean ± SEM; n = 7–12 patients/group; P values are shown (ANOVA with Dunnett’s test). ****P < 0.0001. (F) Schematic of experiment to analyze the effect of NK depletion on anti-TIGIT antibody efficacy. (G) Box-and-whisker plots of percentage of NKp46+NK1.1+ cells in peripheral blood of mice at 2 weeks posttreatment initiation. **P < 0.01. (H) CD45.2+ (mutant) to CD45.1+ (WT) cell ratios and (I) CD11b+Gr1+ percentage in CD45.2+ cells in the peripheral blood of recipient mice. For box-and-whisker plots, boxes represent median, first, and third quartiles, with whiskers extending to 1.5× interquartile range. P values are shown (ANOVA with Dunnett’s test). *P < 0.05; **P < 0.01. ***P < 0.001; ****P < 0.0001. (J) Kaplan-Meier curves of lethally irradiated PepBoyJ CD45.1+ mice receiving whole or NK-depleted CD45.1+ supporting bone marrow cells and 1 × 106 bone marrow cells from Tp53–/–Tet2–/– mice followed by IgG2a isotype control or anti-TIGIT antibody treatment. Log-rank test was used for survival statistics. ****P < 0.0001. Tp53–/–Tet2–/–, Vav-cre Tet2fl/fl Tp53fl/fl; Tet2–/–, Vav-cre Tet2fl/fl; Tp53–/–, Vav-cre Tp53fl/fl; WT, Vav-cre.
We next tested the relevance of upregulation of key immune checkpoint receptors identified above to human disease. We performed spectral flow cytometry on side scatterloCD45dimCD33+ cells from patients with AML with TP53/TET2 comutations (n = 12), TET2 single mutations (n = 9), TP53 single mutations (n = 7), or WT TP53/TET2 (n = 7), separately. CD155 and CD47 were highly expressed on CD33+ AML cells (Figure 9, D and E, and Supplemental Figure 11, B–D). Malignant cells from patients with AML with combined TET2 and TP53 mutations had profoundly elevated CD155 expression compared with other groups (Figure 9, D and E; Supplemental Figure 11, B–D; and Supplemental Table 2). CD155 is a receptor for TIGIT on NK or NKT cells and a marker of CD8+ T cell dysfunction. CD112, also known as Nectin-2 or PVRL2, is an adhesion molecule of the Ig gene superfamily that plays a dual role in immune regulation. CD112 can costimulate T cell responses through CD226, while its interaction with inhibitory receptors like CD112R, TIGIT, and PVRIG suppresses T cell activation. The checkpoint receptor PVRIG competes with DNAM-1 for CD112 binding, and blocking PVRIG-CD112 interactions enhances T cell activation. In NK cells, CD112’s interaction with DNAM-1 promotes cytotoxicity, whereas its binding to TIGIT and PVRIG inhibits NK cell–mediated responses. In addition, TIGIT was upregulated in CD56dimCD16+ and CD56hi NK cells in TP53/TET2 comutant patient AML (Supplemental Figure 12, A–F).
We therefore hypothesized that elevated expression of CD112 and CD155 on AML blasts may contribute to NK or NKT immune evasion in the TET2/TP53 double-mutant microenvironment. To evaluate the effect of the CD155-TIGIT interaction blockade on NK function, we cocultured murine NK cells with leukemic cells from Vav-cre Tet2fl/fl Tp53fl/fl mice in the presence or absence of anti-TIGIT antibody (Supplemental Figure 12G). Anti-TIGIT antibody augmented the ability of NK cells to kill Tp53/Tet2 double-mutant AML. As a control, the isolated NK cells lysed prototypic tumor cell target Yac-1 cells at high efficiency (31). We next treated mice engrafted with Vav-cre Tet2fl/fl Tp53fl/fl AML cells with anti-TIGIT antibody or control. In this aggressive model, CD155 blockade significantly increased mouse survival and substantially reduced malignant cell burden in the animals receiving anti-TIGIT antibody (Supplemental Figure 12, H and I). Additionally, in anti-TIGIT antibody-treated spleens, there were more NK (CD3e–NK1.1+NKp46+) cells within the CD45.1+ tumor immune environment and concomitantly greater NK cell activation (as indicated by granzyme B expression) following anti-TIGIT treatment (Supplemental Figure 12, J and K).
We next determined the requirement of NK cells in response to anti-TIGIT antibody treatment (Figure 9F). Depletion of NK cells mitigated anti-TIGIT–mediated tumor suppression and attenuated anti-TIGIT–mediated inhibition of myeloid bias (Figure 9, G–I). In addition, anti-TIGIT–treated mice with NK depletion experienced anemia, thrombocytopenia, and leukocytosis comparable to isotype control-treated mice (Supplemental Figure 12, L and M). NK depletion completely abrogated the impact of anti-TIGIT antibody treatment on mouse survival (Figure 9J).
Interestingly, CD155 expression was also reduced on Vav-cre Tet2fl/fl Tp53fl/fl cells upon A20 depletion (Supplemental Figure 13, A and B). In parallel to the above, we engrafted CD45.2+ Vav-cre Tet2fl/fl Tp53fl/fl mouse AML cells, CRISPR edited with A20-targeting sgRNAs (sgA20#1 or sgA20#2) or nontargeting control sgRNAs, into lethally irradiated CD45.1+ recipient mice with supporting CD45.1+ bone marrow cells (Supplemental Figure 13C). Similarly to the in vitro results, A20 deletion reduced leukemia burden in vivo (Supplemental Figure 13D). Given that A20 deletion reduced CD155 expression, we tested the effect of anti-TIGIT antibody treatment in this model. A20 deletion resulted in even further reduction in Vav-cre Tet2fl/fl Tp53fl/fl leukemia burden and myeloid bias when coadministered with anti-TIGIT antibody (Supplemental Figure 13, D and E). Anti-TIGIT antibody treatment and A20 KO cooperatively extended the survival time of engrafted mice (Supplemental Figure 13F). These results reveal that AML-mediated NK dysfunction through CD155/TIGIT engagement contributes to AML driven by complete loss of Tp53 and Tet2.
Altogether, these findings demonstrate that combined loss of TP53 and TET2 is associated with profound changes to the immune microenvironment, including increased expansion of monocytic MDSC-like cells, which suppress T cell activation in addition to driving T cell exhaustion and TIGIT-mediated evasion of NK cell–mediated killing.
-
Discussion
Here, we show that TP53 and TET2 mutations cooperate to transform hematopoietic progenitors and play a role in development of AML. As such, these mice provide what we believe to be novel models of AML marked by clonal hematopoiesis and myeloid mutations. We found that hematopoietic progenitor cells in these models were marked by increased activation of innate immune signaling, and exposure of these mice to further environmental proinflammatory signals through TLR2 promoted noncanonical NF-κB pathway activation. These data support previous reports showing that aberrant TLR signaling in early hematopoietic progenitors is associated with a high risk of AML transformation (32) and that increased expression of TLR2 and signaling intermediates (like MyD88) is also seen in the bone marrow of patients with AML with no response to induction therapies compared with those that experience complete remission (32). This study utilized a mouse model with complete loss of Tp53 and Tet2 and therefore may not be representative of all forms of TP53 mutations encountered in AML. As such it was important to evaluate many of the key findings from mice in patient samples with a diverse spectrum of TP53 alterations seen in human AML.
The cooperative impact of TP53 and TET2 mutations in driving AML underscores the importance of understanding the temporal sequence of these mutations. Clonal hematopoiesis often arises from the acquisition of mutations in TET2, which may precede mutations in TP53. This sequence of events may create a proinflammatory environment that promotes the expansion of premalignant clones and transformation to AML. Conversely, TP53 mutations occurring prior to TET2 mutations could result in increased genomic instability and enhanced proliferation, with newly gained TET2 mutations mediating a switch of lineage from lymphoid- to myeloid-biased leukemia. Future studies could leverage inducible mouse models to precisely control the timing and sequence of Tp53 and Tet2 mutations and provide further insight.
TNFAIP3, which encodes the A20 deubiquitinase that activates noncanonical NF-κB signaling, is frequently lost in lymphoma (33) where it promotes lymphomagenesis (33, 34). However, the role of A20 in AML has been less studied (35). A20 KO has been shown to decrease the competitive advantage of cKIT+ bone marrow cells in the TLR-TRAF6–overexpressing MDS mouse model (18). Consistent with these studies, we observed upregulation of A20 in response to TLR2 stimulation and a requirement for A20 in maintenance of TP53/TET2 comutant AML. These data suggest that A20 activity may play roles in proinflammatory signaling and leukemic transformation in TP53/TET2-mutant AML in response to TLR2 signaling.
Beyond tumor-intrinsic effects, we found that TP53 and TET2 comutant AML in both patients and mice exists in an adaptive immune desert environment with reduced abundance of T, B, and NK cells. Moreover, T cells were exhausted in both mouse and human TP53/TET2-mutant AML, and there was clear accumulation of monocytic MDSCs that further suppressed T cell functions in Vav-cre Tet2fl/fl Tp53fl/fl mice. It will be interesting to explore the therapeutic potential of depleting monocytic MDSCs in the future, particularly in relation to their role in the maintenance of AML.
Upregulation of the immune checkpoints CD155 and CD47 was observed on cKit+ malignant cells from Vav-cre Tet2fl/fl Tp53fl/fl mice and in myeloid malignant cells from patients with TP53/TET2 double-mutant AML. Moreover, A20 deletion downregulated the cell surface level of CD155. It is known that CD155 undergoes SUMOylation and inhibition of the SUMO pathway promotes CD155 translocation to the cell surface (36, 37). Therefore, it is possible that A20-mediated CD155 upregulation is dependent on A20-induced deubiquitination or indirect suppression of CD155 degradation. The relationship between A20 and CD155 warrants further investigation.
Upregulation of immune inhibitory checkpoints PD1, CTLA4, and CD47 has been consistently documented in myeloid neoplasms driving many prior clinical trials of antibodies aimed at blocking these proteins in patients with MDS and AML (38–42). Unfortunately, these studies have not yet yielded marked clinical benefits. Excitingly, here we propose a potential therapeutic strategy targeting TIGIT for the treatment of high-risk AML. CD155 interacts with TIGIT, an inhibitory receptor mainly expressed on NK, CD8+ T, and CD4+ T cells, thereby inhibiting the function of T and NK cells (43). Conversely, DNAM1 competes with TIGIT to engage with CD155 to activate NK cells. In support of this notion, CD155 has been reported as a negative prognostic marker for AML (44). We found that blocking the CD155-TIGIT interaction promoted NK cell–mediated killing of AML blasts and conferred survival benefit in AML-engrafted mouse models. We hope these findings will motivate future studies targeting the DNAM1 axis therapeutically in high-risk patients with AML who, based on our data, may likely benefit from TIGIT and CD155 blockade. Our study found overexpression of CD47 in double-KO mice and double-mutant patients, suggesting CD47’s role in immune evasion and leukemic cell survival in this genetic context. Despite CD47 being a promising therapeutic target, clinical trials for AML have shown mixed results. Magrolimab, a monoclonal antibody targeting CD47, demonstrated high response rates in phase Ib trials but was discontinued after the phase III ENHANCE-2 trial owing to lack of survival benefit and anemia-related toxicity (45–47). Other CD47-targeting agents like lemzoparlimab and evorpacept have been explored, with the former’s trials halted and the latter still in development (9, 48). Bispecific therapies like TG1801 showed initial promise but face limited development (48). These results raise questions about CD47 inhibition’s broader applicability in AML. However, it is possible that specific genetic subsets of TP53-mutant AML, such as those with concurrent TET2 mutations, may still derive benefit from CD47-targeted therapies, particularly when combined with other immune-modulating strategies, such as TIGIT blockade. Future studies could explore the functional consequences of CD47 overexpression in this context and evaluate combinatorial approaches targeting multiple immune checkpoints.
-
Methods
Sex as a biological variable. Both sexes were included in clinical sample analyses and animal studies to ensure comprehensive representation. The findings are anticipated to be applicable to both sexes.
Patients. Within the Alliance patient cohort of 1,603 adults and a University of Chicago cohort of 653 adults diagnosed with de novo AML, we identified 315 patients with TP53 mutations, including 128 patients aged <60 years and 187 patients aged ≥60 years, who were treated on the Cancer and Leukemia Group B (CALGB)/Alliance frontline and University of Chicago treatment protocols between 2014 and 2021. Patients with acute promyelocytic leukemia were excluded. Patients were similarly treated with intensive cytarabine/daunorubicin-based induction chemotherapy and consolidation with high-dose chemotherapy or autologous HSC transplantation. Per study protocols, no patient received an allogeneic HSCT in first complete remission. All patients were enrolled on the CALGB 8461 (cytogenetic studies, clinical trial NCT00048958), CALGB 9665 (leukemia tissue bank, clinical trial NCT00899223), and CALGB 20202 (molecular studies) companion protocols.
Mice. Floxed homozygous Tp53 (B6.129P2-Trp53tm1Brn/J, herein called Tp53fl/fl) mice were purchased from The Jackson Laboratory. Vav-cre Tet2fl/fl mice were donated by Ross Levine (Human Oncology and Pathogenesis Program, MSKCC, New York, New York, USA) (16). Peripheral blood was collected by retro-orbital bleeding in EDTA tube, and complete blood count analysis was performed using a Sysmex SN series analyzer (Sysmex Corporation) followed by differential counting. Mice were anesthetized with isoflurane and sacrificed by cervical dislocation upon meeting early removal criteria (e.g., weight loss, hind limb paralysis, lethargy, hunched posture, and difficulty breathing).
Statistics. Statistical analyses were performed using Prism 9.0 software. Mann-Whitney U test was performed between 2 nonparametric groups. For survival analysis, log-rank tests were used. Comparison of multiple groups was evaluated with 1-way ANOVA analysis followed by Dunnett’s test for multiple comparisons. P values of less than 0.05 were considered statistically significant.
Study approval. All procedures performed in the study followed the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and were approved by the Committee on Ethics of Animal Experiments of The Ohio State University and Memorial Sloan Kettering Cancer Center. All patients provided study-specific written informed consent to participate in studies. Studies were performed in accordance with the Declaration of Helsinki and approved by institutional review boards of The Ohio State University, Memorial Sloan Kettering Cancer Center, and the University of Pennsylvania. All patient samples were collected in a pathologically annotated and deidentified fashion according to the guidelines of the ethics boards of each institute and center.
Data availability. Data are available in the Gene Expression Omnibus (GEO GSE217867, GSE285355, and GSE279386). Analysis code is publicly available at https://github.com/blaserlab/lapalombella_pu Values for all data points in graphs are reported in the Supporting Data Values file. Additional methods are provided in the Supplemental Methods.
-
Author contributions
PZ, OAW, BWB, and RL conceptualized the study. PZ, MJC, SDB, OAW, BWB, and RL designed the study methodology. PZ, ECW, SJS, MG, CS, SAS, MC, XP, EDE, TL, BKH, WKC, Y Youssef, BC, AP, AML, CRC, NF, AC, JLL, SD, TK, ASM, DC, HP, KEW, JS, AL, TD, GL, CM, CJW, JSB, AFD, LA, RAB, Y. Yang, NRG, MJC, SDB, BWB, OAW, and RL performed experiments and analyzed data. PZ, AFD, OAW, BWB, and RL provided resources. PZ, OAW, BWB, and RL curated data. PZ, OAW, BWB, and RL wrote the original draft. PZ, OAW, BWB, and RL reviewed and edited the manuscript. PZ, OAW, BWB, and RL acquired funding. OAW, BWB, and RL supervised the study.
-
Acknowledgments
We thank John Byrd for advice on the initial studies, facilitating interaction with the Bloomfield Center, and reviewing the manuscript draft. We also thank Ann-Kathrin Eisfeld and the Bloomfield Center for providing clinical data. The authors are grateful to the patients with AML who provided samples for the above studies and to The Ohio State University Comprehensive Cancer Center (OSUCCC) Leukemia Tissue Bank (supported by NCIP30CA016058 from Ohio State University) for sample procurement. The authors thank OSUCCC Genomics Shared Resource for RNA-Seq. This work was supported by the Damon Runyon Cancer Foundation (to PZ), NIH grant DK128238 (to BWB), National Cancer Institute (NCI; grant P30CA016058), and OSUCCC Pelotonia Foundation funds. OAW is supported by the Edward P. Evans Foundation, NIH/NCI (R01 CA251138, R01 CA242020, P50 CA254838, and R01 CA283364), NIH/National Heart, Lung, and Blood Institute (R01 HL128239), and the Leukemia & Lymphoma Society. We acknowledge the use of the Integrated Genomics Operation Core, supported by the NCI Cancer Center Support Grant (P30 CA08748).
Address correspondence to: Rosa Lapalombella, The Ohio State University, Room 455C, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.6919; Email: rosa.lapalombella@osumc.edu. Or to: Omar Abdel-Wahab, Molecular Pharmacology Program, Sloan Kettering Institute, New York, New York, 10065, USA. Phone: 347.821.1768; Email: abdelwao@mskcc.org. Or to: Bradley W. Blaser, The Ohio State University, Room 302A, OSUCCC Building, 410 West 12th Avenue, Columbus, Ohio, 43210, USA. Phone: 614.685.2341; Email: bradley.blaser@osumc.edu.
-
Footnotes
Conflict of interest: OAW is a founder of and scientific advisor for Codify Therapeutics, holds equity in the company, and receives research funding from it. OAW has served as a consultant for Foundation Medicine Inc., Merck, Prelude Therapeutics, Amphista Therapeutics, MagnetBio, and Janssen and is on the scientific advisory board of Envisagenics Inc., Harmonic Discovery Inc., and Pfizer Boulder. OAW has received prior research funding from H3B Biomedicine, Nurix Therapeutics, Minovia Therapeutics, and LOXO Oncology.
Copyright: © 2025, Zhang et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2025;135(10):e184021.https://doi.org/10.1172/JCI184021.
-
References
- Döhner H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140(12):1345–1377.
- Liu Y, et al. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature. 2016;531(7595):471–475.
- Chen SS, et al. Mutant p53 drives clonal hematopoiesis through modulating epigenetic pathway. Nat Commun. 2019;10(1):5649.
- Loizou E, et al. A gain-of-function p53-mutant oncogene promotes cell fate plasticity and myeloid leukemia through the pluripotency factor FOXH1. Cancer Discov. 2019;9(7):962–979.
- Boettcher S, et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science. 2019;365(6453):599–604.
- Haase D, et al. TP53 mutation status divides myelodysplastic syndromes with complex karyotypes into distinct prognostic subgroups. Leukemia. 2019;33(7):1747–1758.
- Cluzeau T, et al. Phase I/II clinical trial evaluating azacitidine plus venetoclax plus donor lymphocyte infusion in post-transplant relapse myelodysplastic syndromes and acute myeloid leukemia: preliminary results of ventograft, a GFM Study. Blood. 2023;142(suppl 1):3246. View this article via: CrossRef Google Scholar
- Kim M, et al. Mutation in TET2 or TP53 predicts poor survival in patients with myelodysplastic syndrome receiving hypomethylating treatment or stem cell transplantation. Bone Marrow Transpl. 2015;50(8):1132–1134.
- Daver NG, et al. TP53-mutated myelodysplastic syndrome and acute myeloid leukemia: biology, current therapy, and future directions. Cancer Discov. 2022;12(11):2516–2529.
- Tyner JW, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562(7728):526–531.
- Rodriguez-Meira A, et al. Single-cell multi-omics identifies chronic inflammation as a driver of TP53-mutant leukemic evolution. Nat Genet. 2023;55(9):1531–1541.
- Consortium APG. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7(8):818–831.
- Bernard E, et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med. 2020;26(10):1549–1556.
- Shih AH, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell. 2015;27(4):502–515.
- Zhao Z, et al. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev. 2010;24(13):1389–1402.
- Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24.
- Cimmino L, et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell. 2017;170(6):1079–1095.
- Muto T, et al. Adaptive response to inflammation contributes to sustained myelopoiesis and confers a competitive advantage in myelodysplastic syndrome HSCs. Nat Immunol. 2020;21(5):535–545.
- Meisel M, et al. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature. 2018;557(7706):580–584.
- Yeaton A, et al. The impact of inflammation-induced tumor plasticity during myeloid transformation. Cancer Discov. 2022;12(10):2392–2413.
- Tikhonova AN, et al. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569(7755):222–228.
- Wang B, et al. Comprehensive characterization of IFNγ signaling in acute myeloid leukemia reveals prognostic and therapeutic strategies. Nat Commun. 2024;15(1):1821.
- Grieshaber-Bouyer R, et al. The neutrotime transcriptional signature defines a single continuum of neutrophils across biological compartments. Nat Commun. 2021;12(1):2856.
- Jain IH, et al. Genetic screen for cell fitness in high or low oxygen highlights mitochondrial and lipid metabolism. Cell. 2020;181(3):716–727.
- Ramalho-Santos M, et al. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298(5593):597–600.
- Sallman DA, et al. TP53 mutations in myelodysplastic syndromes and secondary AML confer an immunosuppressive phenotype. Blood. 2020;136(24):2812–2823.
- Ianevski A, et al. Fully-automated and ultra-fast cell-type identification using specific marker combinations from single-cell transcriptomic data. Nat Commun. 2022;13(1):1246.
- Khan O, et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature. 2019;571(7764):211–218.
- Xie M, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–1478.
- Scott AC, et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 2019;571(7764):270–274.
- Trotta R, et al. Overexpression of miR-155 causes expansion, arrest in terminal differentiation and functional activation of mouse natural killer cells. Blood. 2013;121(16):3126–3134.
- Monlish DA, et al. The role of toll-like receptors in hematopoietic malignancies. Front Immunol. 2016;7:390.
- Schmitz R, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206(5):981–989.
- Honma K, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood. 2009;114(12):2467–2475.
- Culver-Cochran AE, et al. Chemotherapy resistance in acute myeloid leukemia is mediated by A20 suppression of spontaneous necroptosis. Nat Commun. 2024;15(1):9189.
- Liu J, et al. Targeting the ubiquitination/deubiquitination process to regulate immune checkpoint pathways. Signal Transduct Target Ther. 2021;6(1):28.
- Zitti B, et al. Innate immune activating ligand SUMOylation affects tumor cell recognition by NK cells. Sci Rep. 2017;7(1):10445.
- Abaza Y, Zeidan AM. Immune checkpoint inhibition in acute myeloid leukemia and myelodysplastic syndromes. Cells. 2022;11(14):2249.
- Bewersdorf JP, Abdel-Wahab O. Translating recent advances in the pathogenesis of acute myeloid leukemia to the clinic. Genes Dev. 2022;36(5-6):259–277.
- Haddad F, Daver N. Targeting CD47/SIRPα in acute myeloid leukemia and myelodysplastic syndrome: preclinical and clinical developments of magrolimab. J Immunother Precis Oncol. 2021;4(2):67–71.
- Man CH, et al. Inhibition of PLK4 remodels histone methylation and activates the immune response via the cGAS-STING pathway in TP53-mutated AML. Blood. 2023;142(23):2002–2015.
- Majeti R, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138(2):286–299.
- Chauvin JM, Zarour HM. TIGIT in cancer immunotherapy. J Immunother Cancer. 2020;8(2):e000957.
- Stamm H, et al. Immune checkpoints PVR and PVRL2 are prognostic markers in AML and their blockade represents a new therapeutic option. Oncogene. 2018;37(39):5269–5280.
- Sallman DA, et al. Magrolimab in combination with azacitidine in patients with higher-risk myelodysplastic syndromes: final results of a phase Ib study. J Clin Oncol. 2023;41(15):2815–2826.
- Zeidner JF, et al. Magrolimab plus azacitidine vs physician’s choice for untreated TP53-mutated acute myeloid leukemia: the ENHANCE-2 study. Blood. 2025;:blood.2024027408.
- Daver NG, et al. Tolerability and efficacy of the anticluster of differentiation 47 antibody magrolimab combined with azacitidine in patients with previously untreated AML: phase Ib results. J Clin Oncol. 2023;41(31):4893–4904.
- Wilde L, Kasner M. Targeting CD47: many misses; hopeful for a hit. Blood. 2025;145(5):460–462.
- Traag VA, et al. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep. 2019;9(1):5233.
-
Version history
- Version 1 (March 20, 2025): In-Press Preview
- Version 2 (May 15, 2025): Electronic publication



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)










