Citation Information: J Clin Invest. 2005;115(11):3015-3025. https://doi.org/10.1172/JCI24862.
Abstract
The molecular anatomy of cancer cells is being explored through unbiased approaches aimed at the identification of cancer-specific transcriptional signatures. An alternative biased approach is exploitation of molecular tools capable of inducing cellular transformation. Transcriptional signatures thus identified can be readily validated in real cancers and more easily reverse-engineered into signaling pathways, given preexisting molecular knowledge. We exploited the ability of the adenovirus early region 1 A protein (E1A) oncogene to force the reentry into the cell cycle of terminally differentiated cells in order to identify and characterize genes whose expression is upregulated in this process. A subset of these genes was activated through a retinoblastoma protein/E2 viral promoter required factor–independent (pRb/E2F-independent) mechanism and was overexpressed in a fraction of human cancers. Furthermore, this overexpression correlated with tumor progression in colon cancer, and 2 of these genes predicted unfavorable prognosis in breast cancer. A proof of principle biological validation was performed on one of the genes of the signature, skeletal muscle cell reentry-induced (SKIN) gene, a previously undescribed gene. SKIN was found overexpressed in some primary tumors and tumor cell lines and was amplified in a fraction of colon adenocarcinomas. Furthermore, knockdown of SKIN caused selective growth suppression in overexpressing tumor cell lines but not in tumor lines expressing physiological levels of the transcript. Thus, SKIN is a candidate oncogene in human cancer.
Authors
Francesco Nicassio, Fabrizio Bianchi, Maria Capra, Manuela Vecchi, Stefano Confalonieri, Marco Bianchi, Deborah Pajalunga, Marco Crescenzi, Ian Marc Bonapace, Pier Paolo Di Fiore
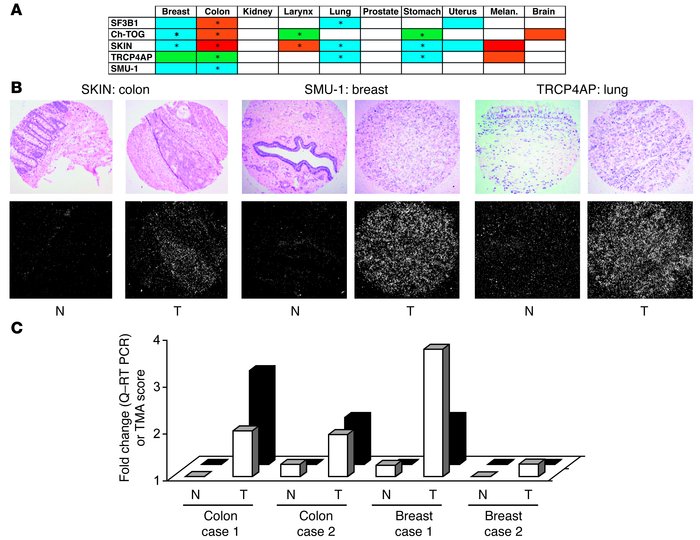
Figure 3



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)