Advertisement
Research ArticleNeuroscienceOncology
Open Access |  10.1172/JCI179395
10.1172/JCI179395
Disruption of ataxia telangiectasia–mutated kinase enhances radiation therapy efficacy in spatially directed diffuse midline glioma models
Avani Mangoli,1 Vennesa Valentine,2 Spencer M. Maingi,2 Sophie R. Wu,2 Harrison Q. Liu,2 Michael Aksu,3 Vaibhav Jain,3 Bronwen E. Foreman,2 Joshua A. Regal,2 Loren B. Weidenhammer,2 Connor E. Stewart,2 Maria E. Guerra Garcia,2 Emily Hocke,2 Karen Abramson,3 Tal Falick Michaeli,1 Nerissa T. Williams,2 Lixia Luo,2 Megan Romero,4 Katherine Deland,2 Samantha Gadd,5 Eita Uchida,5 Laura Attardi,6 Kouki Abe,5 Rintaro Hashizume,5 David M. Ashley,1 Oren J. Becher,4 David G. Kirsch,7 Simon G. Gregory,3 and Zachary J. Reitman2
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Mangoli, A. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Valentine, V. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Maingi, S. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Wu, S. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Liu, H. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Aksu, M. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by
Jain, V.
in:
PubMed
|
Google Scholar
|

1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Foreman, B. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Regal, J. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Weidenhammer, L. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Stewart, C. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Guerra Garcia, M. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Hocke, E. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Abramson, K. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Michaeli, T. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Williams, N. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Luo, L. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Romero, M. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Deland, K. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Gadd, S. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Uchida, E. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Attardi, L. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Abe, K. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by
Hashizume, R.
in:
PubMed
|
Google Scholar
|

1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Ashley, D. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Becher, O. in: PubMed | Google Scholar
1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by
Kirsch, D.
in:
PubMed
|
Google Scholar
|

1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by
Gregory, S.
in:
PubMed
|
Google Scholar
|

1The Preston Robert Tisch Brain Tumor Center,
2Department of Radiation Oncology, and
3The Preston Robert Tisch Brain Tumor Center Omics Program, Duke University, Durham, North Carolina, USA.
4Department of Pediatric Hematology Oncology, Mount Sinai Kravis Children’s Hospital, New York, New York, USA.
5Department of Pediatrics, University of Alabama at Birmingham, Birmingham, Alabama, USA.
6Departments of Radiation Oncology and Genetics, Stanford University School of Medicine, Stanford, California, USA.
7Princess Margaret Cancer Centre, University of Toronto, Toronto, Ontario, Canada.
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
Authorship note: AM and VV are co–first authors.
Find articles by Reitman, Z. in: PubMed | Google Scholar
Authorship note: AM and VV are co–first authors.
Published April 17, 2025 - More info
J Clin Invest. 2025;135(12):e179395. https://doi.org/10.1172/JCI179395.
© 2025 Mangoli et al. This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Received: January 18, 2024; Accepted: April 15, 2025
-
Abstract
Diffuse midline gliomas (DMGs) are lethal brain tumors characterized by p53-inactivating mutations and oncohistone H3.3K27M mutations that rewire the cellular response to genotoxic stress. We used RCAS/tv-a retroviruses and Cre recombinase to inactivate p53 and induce native H3.3K27M mutations in a lineage- and spatially directed manner. We generated primary mouse tumors that recapitulated human DMG. Disrupting ataxia-telangiectasia mutated (ATM) kinase enhanced the efficacy of radiation therapy (RT) in murine and patient-derived DMG models and increased survival. Microscopy-based in situ sequencing was used to spatially resolve transcriptional profiles in more than 750,000 single cells with or without ATM disruption and RT, revealing altered immune-neoplastic and endothelial cell interactions after treatment. An allelic series of primary murine DMG models with different p53 mutations confirmed that transactivation-independent p53 activity was a key mediator of radiosensitivity after ATM disruption. We generated primary DMG mouse models and performed deep profiling that revealed mechanisms of response to ATM disruption and RT that can be utilized as a therapeutic strategy.
-
Introduction
Diffuse midline gliomas (DMGs) are lethal brain tumors that occur in children and young adults. These tumors are localized in essential midline brain structures, such as the brainstem and thalamus, making them surgically inoperable and unresponsive to conventional chemotherapy. The median overall survival of patients with DMGs is less than 2 years. Although radiation therapy (RT) may improve symptoms and extend life, it remains palliative. Somatic activation of lysine 27 to methionine mutations in histone variant 3.3 (H3.3K27M) is a defining feature of DMG (1, 2). Approximately 70% of DMGs harbor inactivating mutations in the tumor suppressor TP53 (1–3) that are associated with radioresistance in patients and preclinical models (4, 5).
A key limitation of current primary DMG preclinical models is the ability to induce K27M mutations in the native H3f3a locus in a spatially, lineage-, and temporally controlled manner. Patient-derived xenografts (6), patient-derived cell lines (7), in utero electroporation (7), and syngeneic mouse models (8, 9) have provided key insights into this disease. A conditional H3f3a-loxP-Stop-loxP-K27M-Tag allele (H3f3aLSL-K27M-Tag) has also been generated that allows the expression of H3.3K27M from the endogenous mouse H3f3a locus in the presence of Cre recombinase (10). However, this model has been limited by cell lineages that can be interrogated with existing Cre driver lines, such as Nestin-Cre (10). To date, the conditional H3.3K27M alleles have not been investigated in an entirely spatially controlled manner. We and others have used the RCAS/tv-a retroviral system for spatially directed modulation of glioma tumorigenesis in mice (5, 11–15). The RCAS/tv-a platform was used to deliver an exogenous H3.3K27M (13, 14, 16), but to our knowledge, it has not been used to edit the endogenous H3f3a allele. A variety of model systems have been used to investigate the mechanisms associated with the development of DMG and to assess therapeutic strategies.
Inhibition of ataxia-telangiectasia mutated (ATM) kinase has emerged as a strategy to enhance the efficacy of RT for DMG (17). ATM is a master orchestrator of the DNA damage response to double-stranded breaks (17). Patients with hereditary loss-of-function ATM variants and those with tumors containing ATM variants are extremely sensitive to RT (17). Consequently, a brain-penetrant ATM inhibitor has entered clinical trials for adult brain tumors (NCT03423628) (18). A recent study identified ATM inhibition as a potent radiosensitization strategy in various patient-derived pediatric high-grade glioma models (6). We found that functional ATM loss–radiosensitized primary mouse models of DMG were driven by p53 loss, but not WT p53 (5, 11). ATM loss increases tumor sensitivity to radiotherapy via radiosensitization of neoplastic cells rather than the vasculature (12). However, it remains uncertain whether H3.3K27M affects the ability of Atm loss to radiosensitize primary DMGs. This is of particular importance, since H3.3K27M regulates the p16 molecular checkpoint that regulates G1-to-S cell-cycle progression (13) and could thereby influence the radiation response.
Here, we examined strategies to exploit the genomically stressed cell state in H3.3K27M/TP53-altered DMG. We improved on previous models that delivered H3.3K27M from an exogenous RCAS payload (11, 13) by combining the RCAS/tv-a system with H3f3aLSL-K27M-Tag mice to express H3.3K27M from the endogenous H3f3a locus. This autochthonous mouse model enabled us to analyze the effect of Atm loss in the context of H3.3K27M/TP53-altered brain tumors to mimic human DMG (10). We found that primary DMGs expressing H3.3K27M driven by p53 loss were radiosensitized by Atm loss. To explore the resistance mechanisms in specific tumor cells, we examined primary mouse DMGs after focal brain irradiation using high-resolution single-cell in situ sequencing (ISS). The results identified the overexpression of the cell-cycle regulator Cdkn1a as a putative resistance factor in Atm-intact DMG. We showed that Cdkn1a, or the transcriptional activity of p53 in general, was dispensable for DMG radiosensitization by Atm loss. Therefore, the nontransactivation functions of p53 may determine the sensitivity of DMGs to combinations of ATM inhibitors and RT. The high-resolution results describe a genetically faithful and flexible primary mouse model of DMG, identifying the mechanisms of resistance to a therapeutic strategy currently in clinical trials.
-
Results
Conditional p53 loss and H3.3K27M expression in retrovirus-induced mouse DMGs. To express H3.3K27M from the endogenous H3f3a locus in retrovirus-induced primary mouse gliomas, we used a H3f3aLSL-K27M-Tag allele that expresses H3.3K27M in the presence of Cre recombinase (10). To incorporate the H3f3aLSL-K27M-Tag allele into the replication-competent avian sarcoma-leukosis virus (ASLV) long terminal repeat (LTR) with splice acceptor (RCAS/tv-a) retrovirus system, mice were bred with NestinTVA mice to allow RCAS retroviruses to specifically transduce TVA+ Nestin–expressing neural stem cells. To investigate the deletion of p53 specific to tumors, we crossbred a p53 variant in which critical exons were flanked by loxP sites (floxed), allowing for functional deletion of p53 in the presence of Cre recombinase. We first introduced retroviruses into NestinTVA p53fl/fl H3f3aLSL-K27M-Tag/+ mice (hereafter referred to as nPH mice) and compared them with matched mice lacking the H3f3aLSL-K27M-Tag allele (hereafter referred to as nP mice) (Figure 1A). We induced DMGs by injecting mice with RCAS retroviruses expressing Cre recombinase, firefly luciferase, and the oncogene platelet-derived growth factor–ligand β (PDGFB) and monitored them for tumor formation by in vivo imaging. Using luciferase-based bioluminescence imaging to detect tumors, we determined that there was no difference in time to tumor formation in H3f3aLSL-K27M-Tag/+ mice compared with matched mice lacking the H3f3aLSL-K27M-Tag allele (Figure 1B). To investigate the effects of Atm deletion in these tumors, we also generated NestinTVA p53fl/fl H3f3aLSL-K27M-Tag/+ Atmfl/fl mice (hereafter referred to as nPHAfl/fl mice) and littermate controls with intact Atm in their tumors of genotype NestinTVA p53fl/fl H3f3aLSL-K27M-Tag/+ Atmfl/+ (hereafter referred to as nPHAfl/+ mice) (Figure 1, C–E, see description of Atm loss results below). Tumors exhibiting hypercellularity and diffuse infiltration of the nearby normal brain on H&E formed within 4–8 weeks with high penetrance (Figure 1F). We detected HA expression indicating the presence of the HA tag on both H3.3K27M and PDGF-β constructs (Figure 1G). As expected, p53 was not detected in p53fl/fl tumors by IHC (Figure 1H). Histone 3 lysine 27 trimethylation was significantly decreased by IHC in H3f3aLSL-K27M-Tag/+ tumors compared with controls (mean, 50.49 % [nP] vs. 5.757 % [nPH] of cells staining positive, P < 0.001; Figure 1I and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI179395DS1), indicating that H3.3K27M could functionally deplete H3K27me3 as predicted (19). Differentially methylated features between nP and K27M-bearing tumors showed hyper- and hypomethylated features within promoters (Supplemental Figure 2A) and enhancers (Supplemental Figure 2B) Additional analysis showed a difference in the percentage of methylation within hypomethylated tiles and de novo tiles in K27M tumors compared with nP and normal murine tissue (Supplemental Figure 2C and D). Differentially methylated genes yielded from the hypomethylated genomic regions were most enriched for processes involving neuronal development and differentiation, suggesting developmental properties for promoter and enhancer tiles (Supplemental Figure 2E). This finding is consistent with the DNA methylation state of other tissues of these specific tiles and with the role of K27M in regulating oncogenic and developmental processes (20). Sequence motif analysis identified differential methylation of motifs associated with the transcription factors Hoxd13 and Hoxa11 (Supplemental Figure 2F), which are known to be involved in hindbrain development. Ki67 was elevated in more than 50% of tumor cells regardless of H3.3K27M status (Figure 1J). Anti-FLAG IHC confirmed the presence of the FLAG tag on the H3.3K27M construct (Figure 1K). FLAG IHC demonstrated that H3.3K27M-Tag+ cells diffusely infiltrated from a hypercellular tumor core into the brain parenchyma, suggesting the diffuse, infiltrative biology seen in human DMG. These results demonstrate that RCAS/tv-a and a conditional H3f3aLSL-K27M-Tag allele can be combined to target K27M to the H3f3a gene in time, lineage, and space to generate primary mouse DMGs that recapitulate human disease.
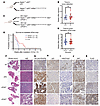 Figure 1
Figure 1Atm loss improves radiosensitivity of primary murine DMBs generated using a conditional H3.3K27M allele. (A) Representation of mouse genotypes used to generate primary mouse DMGs with p53 loss (NestinTVA p53fl/fl [nP]) and mouse DMGs with p53 loss and H3.3K27M (NestinTVA p53fl/fl H3f3aloxP-Stop-loxP-K27M-Tag/+ [nPH]) with or without the conditional H3.3K27M allele. Mice also contained 1 intact and 1 floxed allele of Atm (Atmfl/+, not shown). (B) Dot plot showing the time to tumor formation between nPH and nP mice without any statistical significance. Welch’s t test was used to determine statistical significance. (C) Schematic showing nPHAfl/+ (Atmfl/+) and nPHAfl/fl (Atmfl/fl) within the RCAS/tv-a retrovirus and conditional H3K27M allele. (D) Time to tumor formation showing no statistical difference between nPHAfl/fl and nPHAfl/+ mice. Welch’s t test was used to determine statistical significance. (E) Overall survival of nPHAfl/fl and nPHAfl/+ mice following administration of 3 daily fractions of 10 Gy image-guided focal brain irradiation, with significantly longer median survival of nPHAfl/fl mice. P = 0.03, by Mantel-Cox (log-rank) test. (F) Whole-mount and H&E images of tumor cells from nP, nPH, nPHAfl/+, and nPHAfl/fl (top to bottom) mice exhibiting hypercellularity and infiltration of normal brain tissue. (G) IHC for HA expression indicating the presence of the PDGF-β HA tag. (H) IHC for p53. (I) IHC for histone 3 lysine 27 trimethylation (H3K27me3). (J) IHC displaying the Ki67 proliferation of tumor cells. (K) Anti-FLAG IHC confirmed the presence of the FLAG-HA tag. Scale bars: 100 μm (for all H&E and IHC images in F–K); 1,000 μm (for whole-mount images in F)
Atm loss radiosensitizes primary p53-null/H3.3K27M DMGs. Targeting ATM kinase has emerged as a potential strategy to increase the efficacy of standard-of-care RT for brain tumors (5, 6, 17). We sought to determine whether disruption of ATM could radiosensitize primary mouse DMGs with p53 and H3.3K27M alterations. Previously, we established that H3f3a-WT brainstem gliomas lacking Atm in tumor cells were radiosensitized compared with littermate controls with a functional Atm allele in their tumors (5). However, these mice lack H3.3K27M, which disrupts the G1-to-S cell-cycle checkpoint (13) and may thereby affect the downstream effects of ATM deficiency (17). We hypothesized that Atm inactivation in the presence of the H3.3K27M allele would also radiosensitize tumors. To test this, we assessed the tumor-free survival of nPHAfl/fl mice and compared their survival rates with those of control mice with intact ATM in their tumors of genotype nPHAfl/+ (Figure 1C). We found no difference in tumor-free survival between nPHAfl/fl and nPHAfl/+ mice in the absence of irradiation (Figure 1D). To test whether Atm deletion radiosensitizes p53-null/H3.3K27M DMGs, we delivered 3 daily fractions of 10 Gy focal brain irradiation to mice using the Small Animal Radiation Research Platform (SARRP). nPHAfl/fl mice had significantly longer median survival than did nPHAfl/+ mice (P = 0.03, Mantel-Cox [log-rank test], Figure 1E). Thus, Atm deletion in tumor cells enhanced the efficacy of focal brain irradiation for primary p53-null/H3.3K27M DMGs. H&E staining confirmed tumor cell presence and infiltration (Figure 1F), followed by IHC confirmation of HA expression (Figure 1G), p53 loss (Figure 1H), and the presence of H3.3K27M (Figure 1I), Ki67 (Figure 1J), and anti-FLAG antibodies (Figure 1K). These results show that Atm disruption enhanced the efficacy of RT for primary mouse DMGs that had p53 loss and the H3.3K27M mutation.
In situ multiplexed microscopy reveals cell-cycle and semaphorin pathway changes after irradiation and Atm disruption. To explore the mechanisms underlying radiation efficacy and resistance, we performed spatially resolved gene expression analyses of primary mouse DMGs. Our previous work identified key differences in the response to irradiation and Atm loss between the neoplastic and vascular compartments within primary mouse tumors (12). To distinguish compartment-specific changes in gene expression, such as vascular and immune cells in specific regions of the tumor and nontumor brain, we needed to profile expression changes at single-cell resolution and in a spatially resolved manner. To achieve such a resolution, we used the 10x Genomics Xenium ISS platform to profile primary p53-null/H3.3K27M mouse DMGs. We examined DMG-bearing mice treated or not with focal brain irradiation (10 Gy × 3), with or without tumor Atm loss, as depicted in Figure 2A. We examined 5 μm mid-sagittal sections of formalin-fixed, paraffin-embedded (FFPE) tumor-bearing brains. We supplemented 10x Genomics’ standard mouse brain content with a custom panel containing padlock probes, resulting in 298 brain- and DMG-specific mRNA transcript assays (Supplemental Table 1). Individual cells were detected by nuclear DAPI staining, and cell boundaries were defined by in silico segmentation (see Methods). This yielded 790,374 individual cells across the 4 tumor-bearing brains. Next, we clustered cells on the basis of their transcriptional profiles and compared cell-type composition between the samples. Uniform manifold approximation and projection (UMAP) (21) was performed for reduction, projection, and harmony integration of differentiated normal and neoplastic brain cells into 20 and 29 clusters per specimen, respectively (Figure 2B and Supplemental Figure 3). Examination of differentially expressed marker genes in each cluster identified neoplastic and normal cells including GABAergic interneurons marked by Gad1 and Gad2; microglia marked by P2ry12, Lyz2, and C1qa; and endothelial cells marked by Cd34, Fn1, and Adgrl4 (Supplemental Figure 4). We used canonical cell-type markers and label transfer–based methods to collapse cell clusters into 10 cell archetypes (neoplastic, endothelial, neuronal, astrocytic, oligodendrocytic, microglial, T lymphocytic, etc.) that could be directly compared across specimens (Supplemental Table 2 and Supplemental Figure 5). This analysis revealed mass-like tumors with infiltrating edges recapitulating diffuse glioma biology (Figure 2C). Notably, an Atm-null post-irradiation tumor was smaller and involuted, which was suggestive of rapid treatment response. The tumor core, periphery, and nontumor areas were contoured using these data to allow comparisons between matching cell types and locations after irradiation or Atm loss (Figure 2C).
 Figure 2
Figure 2Spatial clustering in primary mouse DMGs treated with focal brain irradiation or tumoral Atm deletion. (A) Schematic of DMG-bearing mice were subjected or not to focal brain irradiation and ISS. All mice were of the genotype NestinTVA p53fl/fl H3f3aloxP-Stop-loxP-K27M-Tag/+ with either Atm-intact (Atmfl/+) or Atm-null (Atmfl/fl) tumors. (B) Harmony integration showing clustering of 4 tumor-bearing mice with the H3f3aloxP-Stop-loxP-K27M-Tag/+ genotype with either Atm-intact (Atmfl/+) or Atm-null (Atmfl/fl) tumors. (C) Spatial clustering of cells into 10 cell archetypes based on label transfer in 4 tumor-bearing mouse brains (bottom color panel), H&E images of whole brain (left), and distribution of cells within normal brain, tumor periphery, and tumor core annotated in bar graph (right). Top row indicates Atm intact with and without irradiation. Bottom row indicates Atm null with and without irradiation. Color legend on the bottom corresponds to individual cell type noted on bar graph.
We used the spatially resolved expression data to identify differentially expressed genes among neoplastic cells within the tumor cores. We first localized the tumor core within the full-brain sagittal sections using the canonical DMG neoplastic cell markers Olig1, Olig2, and Pdgfra (Figure 3A). As expected, we could not detect p53 in the neoplastic cells within the tumor core in the Tp53fl/fl model, whereas low baseline levels could be detected in non-neoplastic cell types (Figure 3B). Similarly, Atm transcripts were nearly undetectable in neoplastic cells from Atm-null tumors (mean fold-change –0.636, P < 0.0001 vs. Atm-intact tumor; Supplemental Figure 6). To identify transcripts that may be differentially expressed after irradiation and/or Atm loss, we interrogated differentially expressed genes in neoplastic cells after focal brain irradiation in Atm intact tumors (Supplemental Table 3) and Atm-null tumors (Supplemental Table 4). Cyclin-dependent kinase 1a (Cdkn1a), which encodes p21, a potent regulator of cell-cycle progression at G1, was the most differentially expressed gene after focal brain irradiation among Atm intact tumors (log-fold change 0.8, P = 0.0001, by Wilcoxon test, Figure 3C). Cdkn1a was still upregulated, albeit to a lesser degree, after focal brain irradiation among Atm-null tumors (log-fold change 0.6, P = 5.46 × 10–8, by Wilcoxon test, Figure 3D). Conversely, transcription factors such as Sox8 and Sox9, which are associated with developmental cell states, were substantially downregulated after irradiation in Atm-intact tumors, whereas Sox2, Sox4, Pdgfra, and Olig2 were associated with early glial differentiation were all substantially downregulated after irradiation in Atm-null tumors. These results identify the differential expression of cell-cycle regulators and cell-fate–regulating transcription factors after irradiation in a primary DMG mouse model.
 Figure 3
Figure 3Differentially expressed genes and neighborhood analysis of primary mouse DMGs with tumoral Atm loss and/or focal irradiation. (A) Spatial identification of tumors by expression of Pdgfra, Olig1, and Olig2 in all conditions (top to bottom): Atm-intact, Atm-intact with irradiation, Atm-null, Atm-null with irradiation. (B) Spatial identification of p53 loss in all tumor conditions: Atm-intact without and with irradiation (top row, left to right). Atm-null without and with irradiation (bottom row, left to right). (C) Key differentially expressed genes in Atm-intact neoplastic tumor cells treated with and without focal brain irradiation. The log2 fold change and P values for all genes are indicated in Supplemental Table 3. (D) Key differentially expressed genes in Atm-null neoplastic tumor cells treated with and without focal brain irradiation. The log2 fold change and P value for all genes are indicated in Supplemental Table 4. (E) Co-occurrence plot of Atm-intact (nPHAfl/+) tumor showing the number compared with the distance of various cell types in relation to neoplastic cells. (F) Co-occurrence plot of Atm-intact (nPHAfl/+) tumor with irradiation showing the number compared with the distance of various cell types in relation to neoplastic cells. (G) Co-occurrence plot of Atm-null (nPHAfl/fl) tumor showing the number compared with the distance of various cell types in relation to neoplastic cells. (H) Co-occurrence plot of Atm-null (nPHAfl/fl) tumor with irradiation showing the number compared with the distance of various cell types in relation to neoplastic cells. Red arrow indicates increased frequency of immune cells compared with neoplastic cells. Color legend for E–H is on the right side panel. OPC, oligodendrocyte precursor cell; TLC, T lymphocyte. Neighborhood enrichment and co-occurrence analyses were conducted on the entire slide. All unlabeled cells were removed for analysis.
Irradiation and Atm loss were associated with changes in the expression of semaphorin genes specifically, semaphorin 6A (Sema6a) and semaphorin 3D (Sema3d) which have been implicated in the tumor cell proliferation and survival in glioma mouse models and in glioblastomas (22, 23). After irradiation in Atm-intact tumors, Sema3d was significantly increased (log-fold change 1.13, P = 0), suggesting that RT may influence proliferation within the neoplastic core. After irradiation of Atm-null tumors, Sema6a was significantly decreased (log-fold change –0.40, P = 6.59 × 10–15). We utilized single-nucleus RNA-Seq (snRNA-Seq) data from additional primary murine models derived via in utero electroporation approaches to validate semaphorin, p21, and endothelial cell interactions in orthogonal models (Supplemental Figure 7) (9). However, our single-cell spatial transcriptomics provided additional mechanistic insight indicating that after radiotherapy, specific semaphorin genes are altered in neoplastic cells that might play a critical role in glioma biology.
Neighborhood analysis shows altered immune-neoplastic interactions after treatment. Targeting ATM combined with irradiation can bridge innate and adaptive immune processes in extracranial cancers (24, 25). This led us to interrogate the spatial relationship between neoplastic cells and the immune microenvironment. We examined whether the proximity between neoplastic cells and normal cells varied across irradiated or Atm-null tumors. Neighborhood analysis quantified the spatial proximity between different cell types and was used to estimate the mean distance between the neoplastic cells and other cell types (Supplemental Figure 8). These data identified an increased proximity of neoplastic cells and immune cells, such as antigen-presenting cells (APCs) and microglia, after Atm loss and after treatment with irradiation, which was especially pronounced in the irradiated Atm-null tumor. Colocalization analysis between neoplastic cells and other cell types confirmed that microglia and APCs were most enriched within 0–500 μm (Figure 3, E–H), and these cell types were most colocalized in the irradiated Atm-null tumor (Figure 3H).
Ligand-receptor analysis reveals endothelial cell communication. Next, cell-cell and cell-ligand-receptor interactions in primary mouse DMGs established that endothelial cells had the highest frequency of interactions (Supplemental Figure 9 and Supplemental Table 5). We evaluated statistically significant ligand-receptor interactions (P < 0.05) among the tumors and identified the interaction between the endothelium, microglia, and neoplastic cells with decreased Col1a2-CD93 receptor interaction after Atm loss and irradiation (Supplemental Figure 9). CD93 plays a role in tumor-associated vasculature (26), and changes in Col1a2 expression have been observed after radiotherapy in other cancers (27). These results provide insight into the changes in endothelial cell interactions after tumor irradiation. After irradiation of Atm-intact tumors, the cell-ligand interaction of Sema3a:NRP2 between neoplastic cells and microglia decreased. This interaction has been noted to affect glioma cell migration (28), implying a potential alteration in migration with irradiation. We observed the opposite effect in Atm-null tumors after irradiation (Supplemental Figure 9). Thus, ligand-receptor analysis of ISS data suggests that glioma-linked collagen and semaphorin interactions can be examined in primary DMG mouse models.
Pharmacologic ATM inhibition deregulates DNA damage responses and improves survival. To validate these findings, we confirmed that pharmacological inhibition of ATM could radiosensitize patient-derived models of DMG. To do so, we tested whether the brain-penetrant ATM inhibitor AZD1390 (18), combined with focal brain irradiation, could similarly improve the survival of a patient-derived xenograft model of the H3.3K27M-mutant and p53-mutant diffuse midline glioma SF8628 (29–32), which lacks a functional ATM mutation (Supplemental Figure 10 and Supplemental Table 6). The combination of AZD1390 and irradiation significantly extended the median survival of mice compared with either treatment alone (Figure 4A). We tested an Atm-intact genetically engineered model with a combination of AZD1930 and irradiation, which led to a trend for extended median survival compared with irradiation alone (median 29 days vs. 10 days, P = 0.1, log-rank test; Supplemental Figure 11). These results confirmed that pharmacologic or genetic targeting of ATM can radiosensitize multiple types of in vivo DMG models.
 Figure 4
Figure 4Pharmacologic inhibition and DNA damage response signaling in primary mouse DMGs with tumoral Atm loss and/or focal irradiation. (A) Overall survival of mice bearing patient-derived SF8628 DMG xenografts were treated with 20 mg/kg AZD1390 for 2 weeks (ATM inhibitor [ATMi], 5 days per week for 2 weeks) and/or focal brain irradiation (RT, 2 Gy for 3 days per week for a total dose of 12 Gy). (B) Western blot of SF8628 followed by AZD1390 treatment with and without RT (0 hours, 1 hour, 3 hours, 6 hours, 24 hours). (C) Gross dissection and images of the brain with tumor were captured. Representative IHC images of tumor cells from NestinTVA p53fl/fl PDGF-β + H3.3K27M + Cre (PKC) mice treated with vehicle (V) or the ATM inhibitor drug (D) AZD1390, with or without 10 Gy irradiation, for the following (top to bottom): H&E, p-KAP1, total KAP1, and γH2AX. Imaged with Zeiss Axio imager. Original magnification, ×20, insets ×40. (D) PDGF-β + H3.3K27M + p53fl/fl (n = 5 per treatment group) cells stained for p-KAP1 revealed increased p-KAP1 expression in samples treated with 1 dose of 10 Gy and showed significance in vehicle-treated samples (PKC + V +/– RT). P = 0.0079, by Mann-Whitney U test. This expression was significantly reduced in drug-treated tumors subjected to RT compared with vehicle-treated tumors subjected to RT, suggesting the ATM inhibitor AZD1390 sensitized the DIPG tumor–bearing mice to irradiation. *P = 0.0159 and **P = 0.0079, by Mann-Whitney U test. (E) PDGF-β + H3.3K27M + p53fl/fl tumor cells stained for total KAP1 show unchanged levels across all treatment groups (n = 5 per treatment group). (F) PDGF-β + H3.3K27M + p53fl/fl tumor cells stained with yH2AX demonstrated increased expression in samples treated with 1 dose of 10 Gy RT when compared with their respective non-RT-treated samples (n = 5 per treatment group). **P = 0.0079, by Mann-Whitney U test for the vehicle-treated groups and *P = 0.0317, by Mann-Whitney U test for drug-treated groups.
We investigated the DNA damage response in the SF8628 line by performing Western blotting in cells treated with AZD1390 with or without irradiation, which showed increased expression of GH2AX up to 24 hours after irradiation (Figure 4B and Supplemental Figure 12). To further interrogate the effects of irradiation after treatment with AZD1390, we treated Nestin-Tva Cre p53fl/fl PDGF-β H3.3K27M mice (13) with vehicle or drug, along with 10 Gy irradiation and harvested mouse brain tumors 1 hour after irradiation. AZD1390 effectively inhibited ATM, as indicated by significantly reduced phosphorylated KAP1 (p-KAP1) expression in the treated group when compared with expression levels in the control group (P = 0.01, Mann-Whitney U test) and increased GH2AX expression (P = 0.03, Mann-Whitney U test) in tumor-bearing mice (Figure 4, C–F). The differential change in GH2AX expression in tumor-bearing mice treated with AZD1390 when compared with RT alone indicated a synergistic effect.
Atm radiosensitizes Cdkn1a-null primary murine DMGs. Next, we dissected the specific functions of p53 that may affect the radiosensitivity of mouse DMG. Our primary models of DMG indicated that the presence of functional p53 was a key determinant of whether tumors were radiosensitized by Atm loss. For instance, primary p53-null/H3.3K27M tumors and primary p53-null/H3f3a-WT tumors were radiosensitized by Atm loss (Figure 1E and ref. 5, respectively). Conversely, p53 WT primary DMG models driven by inhibitor of cyclin-dependent kinase 4a (Ink4A) and alternative reading frame (ARF) or phosphatase and tensin homolog (PTEN) loss were not radiosensitized by Atm loss (5, 11). However, it is unknown whether the loss of p53 transcriptional activation and/or loss of other p53 functions enables radiosensitization by Atm loss. Our ISS data identified increased Cdkn1a expression in neoplastic cells after radiation. Since Cdkn1a (encoding p21) is a major transcriptional target of p53 (33), we hypothesized that the loss of Cdkn1a function downstream of p53 may be a key determinant of whether Atm loss can radiosensitize primary DMGs. To test whether Cdkn1a loss allows primary mouse DMGs to be radiosensitized by Atm loss, we examined our model of WT p53 DMGs driven by Ink4A/ARF loss, which was not radiosensitized by Atm loss (NestinTVA Ink4A/ARFfl/fl) (5). To test whether p21 loss could radiosensitize these mice when Atm was lost, we bred mice with constitutive p21 loss into this genotype (NestinTVA p21–/– Ink4A/ARFfl/fl Atmfl/fl, referred to hereafter as nlp21Afl/fl mice). We tested the effects of tumor-specific Atm loss by comparing these mice with their littermate controls with intact Atm with the genotype NestinTVA p21–/– Ink4A/ARFfl/fl Atmfl/fl (nIp21Afl/+) (Figure 5A). The time to tumor formation was similar regardless of the presence of intact Atm (Figure 5B). Surprisingly, p21-null mice bearing tumors with Atm deletion had shorter survival following fractionated focal brain irradiation compared with their littermate controls with intact Atm in the tumors (P < 0.03, by log-rank test; Figure 5C). We confirmed p21 loss using IHC (Figure 5, D–E). We found that p21-null tumors with and without Atm loss had similar proliferation indices, as assessed by Ki67 staining (Figure 5F). TUNEL staining of irradiated tumors showed that Atm loss was associated with significantly increased TUNEL staining (P < 0.05, Figure 5, G and H), suggesting that tumors lacking both Atm and Cdkn1a were primed for apoptosis. These results show that functional Cdkn1a was not a key mediator of radiosensitization by Atm loss in the primary mouse model of DMG.
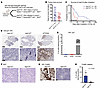 Figure 5
Figure 5Effect of tumor-specific Atm loss in primary DMGs on a Cdkn1a-null (p21–/–)background. (A) Overview of the p21–/– genotypes analyzed. (B) Tumor-free survival of nIp21A mice with and without intact Atm using the log-rank test. (C) Post-focal brain irradiation survival of nIp21A mice with and without intact Atm indicating a statistically significant survival benefit in nlp21Afl/+ mice (P < 0.05, by log-rank test). (D) IHC showing p21 expression in nIp21A mouse brains. NestinTVA Ptenfl/fl Atmfl/+ (nPtenA) tumor–bearing brain generated with identical RCAS viruses shown as a control. (E) Plot indicating the percentage of tumor cells that stained positive for p21 compared with the total cell count. (F) IHC images of staining for Ki67 showing proliferation for nlp21Afl/+ and nIp21Afl/fl. (G) TUNEL staining of tumor-bearing brains of nIp21A mice with and without intact Atm in tumors collected 1 hour after focal brain RT. (H) Quantification of TUNEL staining in nlp21Afl/+ mice. *P < 0.05, by unpaired t test. Scale bars: 2 mm (D top row) 50 μm, (D bottom row, F, and G).
A p53 transactivation domain mutant retains its tumor suppressor function in mouse DMG. Since Atm loss could not radiosensitize Cdkn1a-null DMGs, we reasoned that regulation of p53 transcriptional targets other than Cdkn1a may cause radioresistance in WT p53, Atm-null DMGs. To investigate this possibility, we leveraged the conditional loxP-Stop-loxP-p5325,26 allele (p53LSL-25,26) (34). In the presence of Cre recombinase, this allele expresses p5325,26, a p53 mutant that is severely compromised for the transactivation of most p53 target genes and cannot induce G1 arrest or apoptosis in response to acute DNA damage (34). Interestingly, p5325,26 retains tumor suppressor activity in lung tumors (34), however it is unknown whether it retains tumor suppressor activity in brain tumors. We first determined whether p5325,26 retains tumor suppressor activity in DMG. To test this hypothesis, we compared littermate mice with either p53LSL-25,26/fl or p53fl/fl mice. All mice harbored NestinTVA and were injected with Cre, luciferase, and PDGF-β retrovirus constructs as described above (Figure 6A). We noted a marked delay in tumor presentation in the p53 LSL-25,26/fl group compared with that in the p53fl/fl controls (Figure 6, B and C). Immunohistochemical analysis revealed heterogeneous p53 expression in the p53LSL-25,26/fl group and apparently absent p53 expression in the p53fl/fl group (Figure 6, D and E). Thus, a p53 mutant with severely compromised transactivation activity retained its tumor suppressor activity in primary mouse brainstem gliomas. These results indicate that p53 transactivation function is dispensable for p53 tumor suppression in DMG.
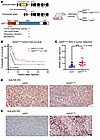 Figure 6
Figure 6Tumor formation in mice expressing a p53 transactivation domain 1 mutant. (A) Schematic for conditional the p53 transactivation domain 1 mutant and mouse genotypes for expression of a p53 transactivation domain 1 mutant. (B) Tumor-free survival in the np53(25,26) group compared with the np53 control group based on a log-rank test. (C) Time to tumor presentation in the p5325,26/fl group compared with the p53fl/fl control group (Wilcoxon test). (D) IHC for anti-HA staining in the p53 and p53(25,26) groups. Scale bars: 50 μm. (E) IHC for p53 expression in the p53 (scale bar: 100 μm) and p53(25,26) (scale bar: 50 μm) groups.
Atm loss does not radiosensitize mouse DMGs lacking a functional p53 transactivation domain. We next sought to determine whether Atm loss could radiosensitize DMGs lacking p53 transcriptional activity but retaining other nontranscriptional functions of p53. We previously showed that Atm loss did not radiosensitize brainstem gliomas driven by Ink4A and ARF loss, however Atm loss modestly radiosensitized brainstem gliomas with both Ink4A and ARF loss and p53 loss (5). We reasoned that if loss of p53 transactivation domain function is the determinant of radiosensitization by Atm loss, then brainstem gliomas with both Ink4A and ARF loss and expression of a transactivation-deficient p5325,26 allele would be radiosensitized by Atm loss. To test if mouse DMGs with p5325,26 and Ink4A and ARF loss were radiosensitized by Atm loss, we bred mice of the NestinTVA p53LSL-25,26/fl Ink4A/ARFfl/fl Atmfl/fl genotype. To test the effects of Atm loss, we compared these mice with their littermate controls with the same genotype except an intact Atm allele (NestinTVA p53LSL-25,26/fl Ink4A/ARFfl/fl Atmfl/+) (Figure 7A). We noted similar time to tumor formation in both models (Figure 7D). Atm loss was associated with differential staining of p-ATM and p-KAP1 after focal brain irradiation (Figure 7, B and C), confirming the loss of ATM functional activity. After subjecting the mice to fractionated focal brain irradiation, no difference in overall survival was appreciated (Figure 7E). These results indicate that the transactivation-independent functions of p53 may be the primary determinants of whether mouse DMGs can be radiosensitized by Atm loss.
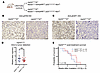 Figure 7
Figure 7Effect of Atm loss on survival after fractionated focal brain irradiation in mouse DMGs expressing a p53 transactivation domain 1 mutant. (A) Schematic showing the p53LSL(25,26) allele and genotypes for NestinTVA p53LSL(25,26)/fl Ink4A/ARFfl/fl mice with either Atmfl/fl or Atmfl/+. (B) IHC images showing p-Atm in Atmfl/+ and Atmfl/fl tumors. Scale bars: 50 μm. (C) IHC images showing p-KAP1 expression in Atmfl/+ and Atmfl/fl tumors. Scale bars: 50 μm. (D) Time to tumor formation in NestinTVA p53LSL(25,26)/fl Ink4A/ARFfl/fl mice with either Atmfl/fl or Atmfl/+ (dot plot). NS, by Wilcoxon test. (E) Overall survival following fractionated brain irradiation in mouse DMGs expressing a p53 transactivation domain 1 mutation with or without Atm loss. The P value is based on a log-rank test.
-
Discussion
Here, we describe the generation of primary mouse DMGs based on recent advances in murine genetic engineering including the conditional H3.3K27M allele and the RCAS/tv-a retrovirus platform (10, 13, 14). We used this model to show that the genetic loss of Atm, an important target for drugs that have entered clinical trials for patients with brain tumors (18), radiosensitizes primary DMG models. Our results in p53-null/H3.3K27M mouse DMGs were similar to those reported previously for p53-null mouse brainstem gliomas (5), in which the sole difference was the presence of H3.3K27M expression from the endogenous H3f3a locus in the neoplastic tumor cells in our current model. In addition, we generated several unique genetically engineered mouse models with differential responses based on genotype (highlighted in Table 1), which suggests that H3.3K27M is not a primary determinant of the ability to target ATM to enhance the efficacy of RT in primary mouse DMG models (Supplemental Figure 13).
 Table 1
Table 1Summary of the effect of Atm loss on radiation sensitivity in genetically engineered DMG mouse models
Our results from genetic experiments in primary mouse models indicate that p53 is a key determinant of the ability of DMGs to be radiosensitized by Atm loss. Almost all p53-altered primary mouse models were radioresistant and radiosensitized by Atm loss, including (a) a model driven by p53 loss with WT H3f3a (5); (b) a model driven by both p53 loss and loss of Ink4A/ARF(5); and (c) the H3.3K27M/TP53 mutant model reported here (Figure 1E). In contrast, Atm loss is unable to radiosensitize primary p53-WT brainstem glioma mouse models, including models driven by Ink4A and ARF loss (5) and models driven by Pten loss (11). Notably, a recent study comprehensively found that pharmacological ATM inhibition radiosensitizes both p53-mutant and WT p53 patient-derived models of DMG and pediatric high-grade glioma (6). Also, H3.3K27M may enhance ATM signaling, increasing radiosensitivity with and without ATM inhibition (35). Together, these data suggest that the mutational status of p53, H3.3K27M, and other alterations should be tested in correlative analyses in future clinical trials of ATM inhibitors in patients with DMG.
Our ISS data provide what we believe to be the first high-resolution transcriptional analysis at high gene plexy (~300 gene targets) in a mouse tumor model, which is critical to defining the model’s tumor vasculature and neoplastic compartments that play distinct roles in the therapeutic response (12). Future work will leverage these data to interrogate tumor immune and vascular microenvironment alterations induced by irradiation and Atm loss, which may guide the rational design of combinations of RT, ATM inhibitors, and therapies targeting the immune system or vasculature.
This work has several limitations. H3.3K27M did not decrease tumor latency in our system, as has been observed in other experimental systems (36). This may be due to the highly restricted manner of H3.3K27M induction in our system (i.e., from the endogeneous H3f3a locus and only in spatially and lineage-restricted cells) and/or the use of a relatively strong PDGF-β co-driver alteration that could mask more subtle H3.3K27M driver phenotypes in our system. Also, the presence of an HA tag on both HA–PDGF-β and H3.3K27M-FLAG-HA constructs precludes specific identification of PDGF-β in the nPH system. Finally, we observed a trend toward improved overall survival with ATM inhibition in an Atm-intact genetic mouse model. We hypothesize that the heterogeneity in tumor latency and the timing of treatment delivery may have conferred a significant survival benefit that was difficult to detect compared with the genetic loss of Atm and compared with the pharmacological xenograft experiment. Future studies could interrogate ATM inhibition effects in Atm-null models to discern on-target versus off-target effects of the ATM inhibitor.
The current work implicates transactivation-independent mechanisms by which p53 mediates radioresistance in Atm-null tumors. Our ISS data show that irradiation elicited overexpression of Cdkn1a, a key downstream target of p53 that mediates cell senescence and G1-to-S checkpoint arrest, in p53-null tumors, indicating p53-independent mechanisms of Cdkn1a expression (37). This finding led us to dissect the contribution of p53 transactivation functions that regulate p21 expression to radiosensitization in Atm-null DMGs. While Atm loss radiosensitizes tumors lacking p53, we found that Atm loss could not radiosensitize tumors containing a p5325,26 allele deficient in p53 transactivation function. Similarly, tumors lacking Cdkn1a (p21) could not be radiosensitized by Atm loss. Our findings highlight the importance of carefully considering p21 status in clinical trials involving Atm inhibition, given the complex role of p21 in tumor growth and the microenvironment (38). Strikingly, we found that Atm loss made tumors more radioresistant in mice that lacked Cdkn1a and that this loss was associated with increased apoptosis. These findings implicate the transactivation-independent function of p53 as a key determinant of radiosensitivity in Atm-null tumors. Future work will dissect the transactivation-independent functions of p53, such as the promotion of apoptosis through mitochondrial membrane permeabilization, direct repression of transcription, and/or direct interaction with complexes that detect DNA lesions (39, 40). Our data provide genetic and mechanistic insights that build upon studies of pharmacological ATM inhibition in patient-derived xenograft models (6). In addition, our work shows that ATM inhibition improves the response to irradiation, leading to extended survival. Further studies are needed to determine the transactivation-independent mechanisms of p53 and ATM-directed therapies and their effect on overcoming resistance to RT in patients with H3.3K27M-mutant DMG.
-
Methods
Sex as a biological variant. Male and female mice were used in all murine experiments to ensure representation of both sexes. We did not identify any sex-specific differences in the data, and all findings were consistent across male and female mice. As such, sex was not considered to be a biological variable in the interpretation of the results. The outcomes of this study are therefore expected to be broadly relevant to both sexes.
Detailed workflows for the generation, brain irradiation, and molecular analysis of primary mouse DMG models using RCAS/tv-a and Cre/loxP technologies are described in our recent publication (41). Male and female mice were utilized for all murine models. All new reagents, materials, and software are listed in and software are listed in Supplemental Table 8. A list of the abbreviations used is provided in Supplemental Table 7.
Mouse strains. Detailed workflows for generation, brain irradiation, and molecular analysis of primary mouse DMG models using RCAS/tv-a and Cre/loxP technologies are found in our recent STAR Protocols publication (41). Complex mouse strains were generated by breeding mice with the following alleles: NesTVA, Atmfl, and p53fl (41), Ink4A/ARFfl (41), p53LSL(25,26) (34), and p21–/– (42). The H3f3aLSL-K27M-Tag allele was a gift from Suzanne Baker, St. Jude Children’s Research Hospital, Memphis Tennessee (10).
DF1 cell culture and retrovirus generation. DF1 cells were cultured in DMEM containing 10% FBS, and RCAS/tv-a retroviruses were generated using RCAS-Cre, RCAS-luc, and RCAS-PDGF-β plasmids as previously described (41).
Mouse brainstem injection. The harvested DF1 cells were injected into the brainstems of mice anesthetized on ice on P3–P5 as previously described (41). Patient-derived xenografts using the SF8628 model were generated by brainstem injection as described before (29–31).
Mouse in vivo imaging. Bioluminescence imaging of gliomas in mice was performed by intraperitoneal injection of d-luciferin and IVIS Illumina III as previously described (41).
Image-guided focal brain irradiation. Irradiation of gliomas in mice was delivered on a SARRP using image-guided, opposed-lateral beams as described previously (41). Daily fractions of 10 Gy were delivered for primary DMG models on 3 consecutive days. For patient-derived xenograft DMG models, a dose of 2 Gy was delivered 3 days a week, for a total dose of 12 Gy.
ATM inhibitor studies. For the patient-derived xenograft model, SF8628 (H3.3K27M DIPG) was obtained from the UCSF Medical Center. Establishment of SF8628 cell culture from surgical specimens and tumor cell modification for expression of firefly luciferase for in vivo bioluminescence imaging have been described in previous publications (30–32). SF8628 cells were propagated as monolayers in complete medium consisting of DMEM supplemented with 10% FBS and nonessential amino acids. Short tandem repeats (STRs) were obtained to confirm the identity of the cell lines. All cells were cultured in an incubator at 37°C in a humidified atmosphere containing 95% O2 and 5% CO2 and were free of mycoplasma at the time of testing. Six-week-old female athymic mice (rnu/rnu genotype, BALB/c background) were purchased from Envigo and housed under aseptic conditions. Pontine injection of tumor cells was performed as previously described (29–31). Each mouse was injected with 1 μL SF8628 cell suspension (100,000 cells/μL) into the pontine tegmentum at a depth of 5 mm from the inner base of the skull. For the efficacy study of AZD1390 and radiation, animals were randomized into 4 treatment groups: (a) vehicle control (0.5% hydroxymethylcellulose, 0.1% Tween 80, n = 6); (b) ADZ1390 monotherapy (oral gavage of 20 mg/kg AZD1390 for 5 times per week for 2 consecutive weeks, n = 6); (c) radiation monotherapy (2.0 Gy, 3 times per week for 2 consecutive weeks for a total dose of 12 Gy, n = 6); and (d) AZD1390 and radiation combination therapy (n = 6). Biweekly bioluminescence imaging was used to monitor tumor growth and response to therapy as previously described (29). Mice were monitored daily and euthanized at the endpoint, which included irreversible neurological deficit or a body condition score of less than 2. For the genetically engineered mouse model, gliomas were generated using the RCAS/Tv-a system as previously described (43, 44). DF1 cells were transfected with RCAS plasmids (RCAS–PDGF-β, RCAS-Luc, and RCAS-Cre, mixed at a 1:1:1 ratio prior to intracranial injections). nP mice were intracranially injected with 1.0 μL RCAS virus producing DF1 cells at P3 to 5 (P3–P5) and monitored 3 times per week after weaning for signs of brain tumor symptoms (enlarged head, ataxia, weight loss up to 20%). Bioluminescence imaging was then used to monitor tumor formation from weeks 4–12 as described previously (38). Following tumor detection via imaging or the onset of neurological symptoms, mice were randomized 1:1 into 2 treatment groups: RT alone or AZD1390 plus RT. AZD1390 was obtained from AstraZeneca and resuspended in 0.5% w/v is hydroxypropyl methylcellulose and 0.1% w/v Tween 80 solution. Mice in the AZD1390 plus RT group were dosed at 20 mg/kg via oral gavage 1 hour prior to RT, as suggested by the superior efficacy dose (18). Both treatment groups received 3 consecutive daily fractions of 10 Gy focal brain irradiation delivered by the SARRP. Treated mice were monitored for survival until they reached the humane endpoint. (14, 18)
IHC. IHC analysis was performed using methods previously described for Ki67, p-Atm, p-KAP1, total KAP1, γH2AX, and p53 (18, 41). Additional IHC and TUNEL staining was performed by HistoWiz (histowiz.com) using a standard operating procedure and a fully automated workflow for p21 (Cdkn1a), FLAG, and TUNEL on a Bond Rx autostainer (Leica Biosystems) with enzyme treatment (1:1,000) using standard protocols. H&E staining was performed using standard protocols.
Xenium in situ and bioinformatics analysis. Tumor-bearing brains subjected to Xenium ISS were detected by in vivo imaging 37–48 days after birth and collected 7 days after tumor detection, either after 3 daily treatments of 10 Gy initiated within 2 days of tumor detection or after mock treatment. Initial data generated by the Xenium instrument (45) are processed on board with a built-in analysis tool called the Xenium Analyzer (45). The Xenium Analyzer is fully automated and includes an imager (imageable area of approximately 12 × 24 mm per slide), sample handling, liquid handling, wide-field epifluorescence imaging, capacity for 2 slides per run, and an on-instrument analysis pipeline. The analysis pipeline included image preprocessing, puncta detection, transcript decoding, and quality score assignment. The pipeline also performed cell segmentation using DAPI images to detect nuclei using a neural network. Each nucleus was then expanded outward until either a maximum distance of 15 μm was reached or the boundary of another cell was reached. A variety of output files were produced using an on-instrument pipeline. The essential files for downstream analysis included the feature-cell matrix (HDF5 and MEX formats identical to those output by single-cell RNA tools from 10x Genomics [Cellranger/Spaceranger]), the transcripts (listing each mRNA, its 3D coordinates, and a quality score), and the cell boundaries CSV file.
Xenium output was first imported into R (4.3.1) using the LoadXenium function from Seurat (version 4.9.9.9050) (46). The 4 Xenium samples were processed using Seurat (46). Data were loaded and filtered using an nFeature_Xenium>5 and nCount_Xenium of greater than 10 as a criterion. Cells without a predicted annotation were then subset out, and the 4 samples were normalized using SCTransform. Principal component analysis was also run for each sample. The 4 samples were then integrated using the IntegrateLayers function, with HarmonyIntegration as the method (47).
Specific regions of the tissue were annotated manually using the polygon tool in Xenium Explorer software (development version, 10x Genomics), and the polygon coordinates were exported as.csv files. Cells with zero counts were then filtered, and the points within the polygon coordinates were identified using the point.in.polygon function in the sp (2.0.0) R package. Additional plots were generated using Seurat, and deconvolution was performed with spacexr (version 2.2.1) (48) using a custom annotated single-cell reference from a previous experiment. Differential gene expression analysis across regions of interest (ROIs) was assessed with the Wilcoxon test on SCTransform normalized count data using the FindMarkers Seurat function. Co-occurrence and neighborhood enrichment plots were generated using the Python package Squidpy (version 1.2.3) (49), and trajectory and cell-cell-interaction analyses were performed using the Python package STLearn (version 0.4.12) (50).
All the cells on the entire slide were used to determine the different types of cell clusters. All unlabeled cells were removed for Squidpy and STLearn analyses. Co-occurrence and neighborhood enrichment analyses were conducted on all cells within the entire slide. The tumor core and periphery were utilized to identify cell-cell and cell-ligand interactions. Differentially expressed genes were analyzed in the tumor core.
Whole-genome bisulfite sequencing and bioinformatics analysis. Bisulfite methylation sequencing and data analysis were performed by Novogene. Briefly, K27M-mutant (nPH) and matched WT K27M (nP) tumor-bearing mice (n = 4 biological replicates per group) were generated as described above, and tumor-bearing brains were embedded in FFPE. Tumor regions were identified in the brains on matched H&E-stained slides, and tumor microdissection was performed. Genomic DNA was isolated, spiked with lambda bacteriophage DNA (to serve as an internal negative control), and fragmented to 200–400 base pairs, and bisulfite treatment was performed to convert unmethylated cytosines into uracil via deamination. Notably, this process does not alter methylated cytosines, allowing identification of these sites downstream. After methylation sequencing adapter ligation, dsDNA synthesis, and library size selection, PCR amplification was performed followed by Illumina sequencing. FastQC was used for quality control on the raw reads. Bismark software (version 0.24.0) was used to perform alignments of bisulfite-treated reads to a reference genome (-X 700--dovetail). Bismark software is a tool utilized for bisulfite sequencing data analysis and the source code is freely available. The reference genome was firstly transformed into a bisulfite-converted version (C-to-T and G-to-A converted) and then indexed using bowtie2, a software tool utilized for aligning sequencing reads to the reference genome. Sequence reads were also transformed into fully bisulfite-converted versions (C-to-T and G-to-A converted) before they were aligned to similarly converted versions of the genome in a directional manner. Sequence reads that produced a unique best alignment from the 2 alignment processes (original top and bottom strand) were then compared with the normal genomic sequence, and the methylation state of all cytosine positions in the read was inferred. The same reads that aligned to the same regions of the genome were regarded as duplicated ones. The sequencing depth and coverage were summarized using deduplicated reads. The results of the methylation extractor (bismark_methylation_extractor,-- no_overlap) were transformed into the bigWig format for visualization using the Integrative Genomics Viewer (IGV) browser. The sodium bisulfite nonconversion rate was calculated as the percentage of cytosine sequenced at cytosine reference positions in the lambda genome. Genes were extracted from genome assembly GRCm39 using Ensembl gene set version 111. Promoter regions were defined from 1,500 bp upstream of the transcription start site (TSS) to 500 bp downstream. For each region, methylated CpG reads and unmethylated reads were counted and summed, and the average methylation level was calculated. Similarly, putative enhancers were extracted from Ensembl regulation version 111 of GRCm39, and an average methylation level was calculated for each enhancer. The methylation difference was determined for each feature (promoter and enhancer) between samples from the K27M group and samples from the nPA group. Significance was estimated by applying ANOVA over a linear mode fit (51). For DNA methylation extraction, the methikit was used. Motif analysis was carried out using HOMER (http://homer.ucsd.edu/homer). For pathway enrichment analysis, STRING (https://string-db.org/) was used. For “other tissues” we analyzed data from the NCBI Gene Expression Omnibus (GEO) database (GSE42836). Methylation extraction was done using the methkit.
Statistics. Statistical significance in volcano plots across ROIs was assessed with the Wilcoxon test on SCTransform-normalized count data using the FindMarkers Seurat function. Data plotting and quantification analyses were performed using GraphPad Prism 9 (GraphPad Software). An unpaired 2-tailed t test, Mann Whitney test, and/or one way ANOVA was utilized to determine significance in IHC quantifications was utilized to determine significance in IHC quantifications. The log-rank test was used to determine the survival rate. The Wilcoxon test was used to determine differences in the time to tumor detection. Individual data points are plotted, and all statistically significant values (P < 0.05) are identified with an asterisk. Data points are presented as the mean with SD.
Study approval. All animal studies were conducted according to institutional animal protocols. All animal experiments were approved by the IACUCs of Duke University and Northwestern University. For the patient-derived xenograft model, the human cell line SF8628 (H3.3K27M DIPG) was obtained from the UCSF Medical Center, in accordance with an institutionally approved protocol.
Data availability. All data were deposited in the NCBI GEO database (GSE246584; for whole-genome bisulfite sequencing, GSE284759). Additional information required to reanalyze the data reported in this work is available from the corresponding author upon request. All raw values for figures are available in the Supporting Data Values file.
-
Author contributions
AM prepared the manuscript, designed experiments, and analyzed data. VV designed and led the execution of experiments for the revised manuscript including mouse tissue analyses, whole-genome bisulfite sequencing, and pharmacologic in vivo experiments. AM and VV share the first author position. The order of the first authors’ names was determined by alphabetical order of the last names (the order was decided to be A–Z on the basis of a coin-flip by the corresponding author ZJR). AM, ZJR, SGG, DGK, DMA and OJB designed the study and experiments. SRW, HQL, BEF, LBW, MEGG, DGK, LL, KD, VV, SMM, MR and ZJR performed mouse experiments and tabulated data. NTW performed mouse irradiation procedures. K Abe, EU, and RH performed patient-derived xenograft and ATM inhibitor pharmacologic experiments. LA contributed the p53 transactivation mutant mouse strain and experimental design regarding the strain. EH, K Abramson, LBW, and ZJR assisted with in situ sequencing experiments. MA, VJ, JAR, SG and ZJR performed bioinformatics analyses. TFM performed whole-genome bisulfite sequencing analyses. CES performed mouse experiments and tabulated data. AM, ZJR, and SGG prepared the manuscript.
-
Acknowledgments
We thank Suzanne Baker (St. Jude Research Hospital, Memphis, Tennessee, USA) for the gift of the H3f3a-loxP-Stop-loxP-K27M-Tag mice. Funds for ISS data generation were provided by the Duke Brain Tumor Omics Program (Durham, North Carolina, USA). This study was supported by a National Cancer Institute (NCI), NIH, Mentored Clinician Scientist Development Award (K08256045); an Alex’s Lemonade Stand Foundation A Award (no. 23-27774); the ChadTough Defeat DIPG Foundation; the SoSo Strong Foundation; the Pediatric Brain Tumor Foundation; the Emily Beazley’s Kures for Kids Fund; the St. Baldrick’s Foundation; the Lauren Brescia Memorial Fund; NCI (P50CA190991) Duke SPORE in Brain Cancer developmental funds (to ZJR); the NCI, NIH (7R35CA197616, to DGK); and Hyundai Hope on Wheels (to OJB).
Address correspondence to: Zachary J. Reitman, Duke University, Department of Radiation, 30 Duke Medicine Circle, Box 3085, Durham, North Carolina 27710, USA. Phone: 919.668.7336; Email: zjr@duke.edu. Or to: Simon G. Gregory, 300 N. Duke Street, DUMC 104775, Durham, North Carolina 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu.
-
Footnotes
Conflict of interest: DGK is a cofounder and stockholder in XRAD Therapeutics, which is developing radiosensitizers. DGK is a member of the scientific advisory board and owns stock in Lumicell Inc., a company commercializing intraoperative imaging technology. DGK is a co-inventor of the Lumicell handheld imaging device (US patent no. 20140301950-A1) and is a co-inventor on a patent for radiosensitizers, Dual ATM and PK Inhibitors for use in anti-tumor therapy (US patent no. 12187742). XRAD Therapeutics, Merck, Bristol Myers Squibb, and Varian Medical Systems have provided research support to DGK. ZJR receives royalties for intellectual property related to brain tumor diagnostic tests that is managed by the Duke Office of Licensing and Ventures and has been licensed to Genetron Health and receives honoraria for teaching from Oakstone Publishing and Eisai Pharmaceuticals.
Copyright: © 2025, Mangoli et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2025;135(12):e179395.https://doi.org/10.1172/JCI179395.
-
References
- Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–253.
- Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231.
- Zhang L, et al. Exome sequencing identifies somatic gain-of-function PPM1D mutations in brainstem gliomas. Nat Genet. 2014;46(7):726–730.
- Werbrouck C, et al. TP53 pathway alterations drive radioresistance in diffuse intrinsic pontine gliomas (DIPG). Clin Cancer Res. 2019;25(22):6788–6800.
- Deland K, et al. Tumor genotype dictates radiosensitization after Atm deletion in primary brainstem glioma models. J Clin Invest. 2021;131(1):e142158.
- Xie J, et al. ATM inhibition enhances the efficacy of radiation across distinct molecular subgroups of pediatric high-grade glioma. Neuro Oncol. 2023;25(10):1828–1841.
- Khadka P, et al. PPM1D mutations are oncogenic drivers of de novo diffuse midline glioma formation. Nat Commun. 2022;13(1):604.
- du Chatinier A, et al. Generation of immunocompetent syngeneic allograft mouse models for pediatric diffuse midline glioma. Neurooncol Adv. 2022;4(1):vdac079.
- McNicholas M, et al. A compendium of syngeneic, transplantable pediatric high-grade glioma models reveals subtype-specific therapeutic vulnerabilities. Cancer Discov. 2023;13(7):1592–1615.
- Larson JD, et al. Proteogenomic characterization of human early-onset gastric cancer. Cancer Cell. 2019;35(1):111–124.
- Stewart CE, et al. The effect of Atm loss on radiosensitivity of a primary mouse model of Pten-deleted brainstem glioma. Cancers (Basel). 2022;14(18):4506.
- Deland K, et al. Radiosensitizing the vasculature of primary brainstem gliomas fails to improve tumor response to radiation therapy. Int J Radiat Oncol Biol Phys. 2022;112(3):771–779.
- Cordero FJ, et al. Histone H3.3K27M represses p16 to accelerate gliomagenesis in a murine model of DIPG. Mol Cancer Res. 2017;15(9):1243–1254.
- Hoeman CM, et al. ACVR1 R206H cooperates with H3.1K27M in promoting diffuse intrinsic pontine glioma pathogenesis. Nat Commun. 2019;10(1):1023.
- Hambardzumyan D, et al. Modeling adult gliomas using RCAS/t-va technology. Transl Oncol. 2009;2(2):89–95.
- Tomita Y, et al. A novel mouse model of diffuse midline glioma initiated in neonatal oligodendrocyte progenitor cells highlights cell-of-origin dependent effects of H3K27M. Glia. 2022;70(9):1681–1698.
- Garcia MEG, et al. Targeting the ATM kinase to enhance the efficacy of radiotherapy and outcomes for cancer patients. Semin Radiat Oncol. 2022;32(1):3–14.
- Durant ST, et al. The brain-penetrant clinical ATM inhibitor AZD1390 radiosensitizes and improves survival of preclinical brain tumor models. Sci Adv. 2018;4(6):eaat1719.
- Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861.
- Filbin MG, et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science. 2018;360(6386):331–335.
- van der Maarten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9(86):2579–2605.
- Zhao J, et al. SEMA6A is a prognostic biomarker in glioblastoma. Tumour Biol. 2015;36(11):8333–8340.
- Angelucci C, et al. Multifaceted functional role of semaphorins in glioblastoma. Int J Mol Sci. 2019;20(9):2144.
- Hu M, et al. ATM inhibition enhances cancer immunotherapy by promoting mtDNA leakage and cGAS/STING activation. J Clin Invest. 2021;131(3):e139333.
- Zhang Q, et al. Inhibition of ATM increases interferon signaling and sensitizes pancreatic cancer to immune checkpoint blockade therapy. Cancer Res. 2019;79(15):3940–3951.
- Tossetta G, et al. Role of CD93 in health and disease. Cells. 2023;12(13):1778.
- Yao G, et al. Radiation increases COL1A1, COL3A1, and COL1A2 expression in breast cancer. Open Med (Wars). 2022;17(1):329–340.
- Nasarre C, et al. Neuropilin-2 acts as a modulator of Sema3A-dependent glioma cell migration. Cell Adh Migr. 2009;3(4):383–389.
- Katagi H, et al. Radiosensitization by histone H3 demethylase inhibition in diffuse intrinsic pontine glioma. Clin Cancer Res. 2019;25(18):5572–5583.
- Hashizume R, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20(12):1394–1396.
- Aoki Y, et al. An experimental xenograft mouse model of diffuse pontine glioma designed for therapeutic testing. J Neurooncol. 2012;108(1):29–35.
- Pal S, et al. Dual HDAC and PI3K inhibition abrogates NFκB- and FOXM1-Mediated DNA damage response to radiosensitize pediatric high-grade gliomas. Cancer Res. 2018;78(14):4007–4021.
- Brugarolas J, et al. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377(6549):552–557.
- Brady CA, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145(4):571–583.
- Peterson E, et al. Inhibition of H3K27M-enhanced ATM signaling increases radiation efficacy in diffuse midline glioma [preprint]. https://doi.org/10.1101/2024.11.01.621526 Posted on bioRxiv November 3, 2024.
- Silveira AB, et al. H3.3 K27M depletion increases differentiation and extends latency of diffuse intrinsic pontine glioma growth in vivo. Acta Neuropathol. 2019;137(4):637–655.
- Galanos P, et al. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat Cell Biol. 2016;18(7):777–789.
- Hukkelhoven E, et al. Tyrosine phosphorylation of the p21 cyclin-dependent kinase inhibitor facilitates the development of proneural glioma. J Biol Chem. 2012;287(46):38523–38530.
- Boutelle AM, Attardi LD. p53 and Tumor Suppression: It Takes a Network. Trends Cell Biol. 2021;31(4):298–310.
- Ho T, et al. How the other half lives: What p53 does when it is not being a transcription factor. Int J Mol Sci. 2019;21(1):13.
- Weidenhammer LB, et al. Inducing primary brainstem gliomas in genetically engineered mice using RCAS/TVA retroviruses and Cre/loxP recombination. STAR Protoc. 2023;4(1):102094.
- Deng C, et al. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82(4):675–684.
- Misuraca KL, et al. Pax3 expression enhances PDGF-B-induced brainstem gliomagenesis and characterizes a subset of brainstem glioma. Acta Neuropathol Commun. 2014;2:134.
- Misuraca KL, et al. A novel mouse model of diffuse intrinsic pontine glioma initiated in Pax3-expressing cells. Neoplasia. 2016;18(1):60–70.
- Janesick A, et al. High resolution mapping of the breast cancer tumor microenvironment using integrated single cell, spatial and in situ analysis of FFPE tissue. Nat Commun. 2023;14(1):8353.
- Hao Y, et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol. 2024;42(2):293–304.
- Korsunsky I, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–1296.
- Cable DM, et al. Cell type-specific inference of differential expression in spatial transcriptomics. Nat Methods. 2022;19(9):1076–1087.
- Palla G, et al. Squidpy: a scalable framework for spatial omics analysis. Nat Methods. 2022;19(2):171–178.
- Pham D, et al. Robust mapping of spatiotemporal trajectories and cell–cell interactions in healthy and diseased tissues. Nat Commun. 2023;14:7739.
- Falick Michaeli T, et al. Muscle injury causes long-term changes in stem-cell DNA methylation. Proc Natl Acad Sci U S A. 2022;119(52):e2212306119.
-
Version history
- Version 1 (April 17, 2025): In-Press Preview
- Version 2 (June 16, 2025): Electronic publication



Copyright © 2026 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)









