Figure 1
Options:
View larger image
(or click on image)
Download as PowerPoint
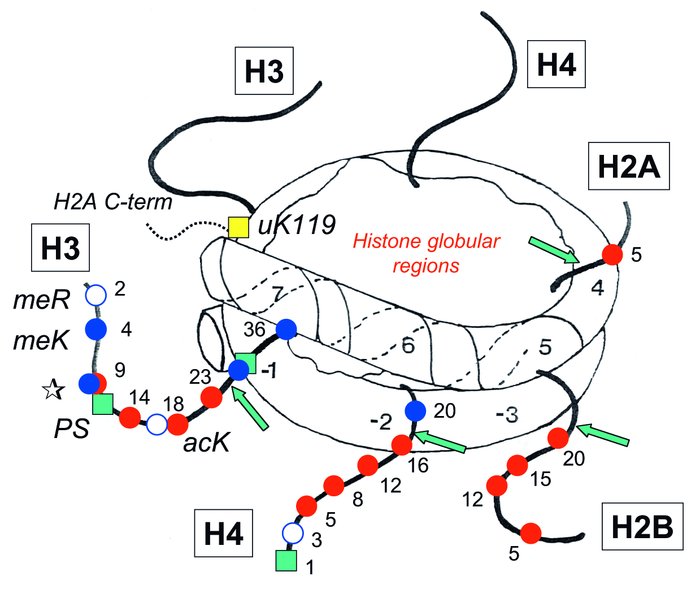
A nucleosome core particle, showing the core histone N-terminal tail domains and sites of posttranslational modification. Residue numbers for modified residues are shown. Note that H3 lysine 9 (star) can be either acetylated or methylated. The C-terminal tail domain of one H2A molecule is shown (dashed line) with the site of ubiquitination at lysine 119 (yellow square). Modifications are shown on only one of the two copies of histones H3 and H4, and only one tail is shown for H2A and H2B. Numbers along the DNA indicate each complete helical turn on either side of the dyad axis. Sites marked by green arrows are susceptible to cutting by trypsin in intact nucleosomes. Red circles, acetyl lysine (acK); blue circles, methyl lysine (meK); white circles, methyl arginine (meR); green squares, phosphoryl serine (PS); yellow square, ubiquitinated lysine (uK).



Copyright © 2026 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)