Abstract
Recent controversies surrounding prostate cancer overtreatment emphasize the critical need to delineate the molecular features associated with progression to lethal metastatic disease. Here, we have used whole-genome sequencing and molecular pathological analyses to characterize the lethal cell clone in a patient who died of prostate cancer. We tracked the evolution of the lethal cell clone from the primary cancer to metastases through samples collected during disease progression and at the time of death. Surprisingly, these analyses revealed that the lethal clone arose from a small, relatively low-grade cancer focus in the primary tumor, and not from the bulk, higher-grade primary cancer or from a lymph node metastasis resected at prostatectomy. Despite being limited to one case, these findings highlight the potential importance of developing and implementing molecular prognostic and predictive markers, such as alterations of tumor suppressor proteins PTEN or p53, to augment current pathological evaluation and delineate clonal heterogeneity. Furthermore, this case illustrates the potential need in precision medicine to longitudinally sample metastatic lesions to capture the evolving constellation of alterations during progression. Similar comprehensive studies of additional prostate cancer cases are warranted to understand the extent to which these issues may challenge prostate cancer clinical management.
Authors
Michael C. Haffner, Timothy Mosbruger, David M. Esopi, Helen Fedor, Christopher M. Heaphy, David A. Walker, Nkosi Adejola, Meltem Gürel, Jessica Hicks, Alan K. Meeker, Marc K. Halushka, Jonathan W. Simons, William B. Isaacs, Angelo M. De Marzo, William G. Nelson, Srinivasan Yegnasubramanian
Figure 1
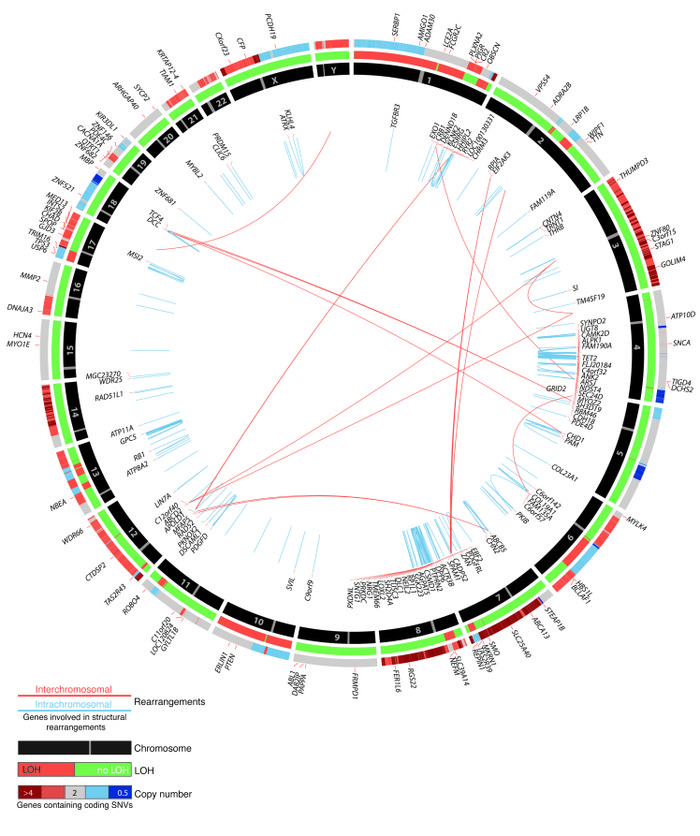
Common genomic consensus alterations found in 3 distant metastases (M5, M38, and M40) by whole-genome sequencing are plotted.



Copyright © 2026 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)