Advertisement
Review
Open Access |  10.1172/JCI191645
10.1172/JCI191645
Biological and clinical implications of a model of surveillance immunity
Katharina Willmann and Luis F. Moita
Center for Disease Mechanisms Research, Faculdade de Medicina da Universidade de Lisboa, Lisbon, Portugal.
Address correspondence to: Luis F. Moita, Faculdade de Medicina, Universidade de Lisboa, School of Medicine, University of Lisbon, Avenida Professor Egas Moniz, 1649-028 Lisboa, Portugal. Email: lmoita@medicina.ulisboa.pt.
Find articles by Willmann, K. in: PubMed | Google Scholar
Center for Disease Mechanisms Research, Faculdade de Medicina da Universidade de Lisboa, Lisbon, Portugal.
Address correspondence to: Luis F. Moita, Faculdade de Medicina, Universidade de Lisboa, School of Medicine, University of Lisbon, Avenida Professor Egas Moniz, 1649-028 Lisboa, Portugal. Email: lmoita@medicina.ulisboa.pt.
Find articles by
Moita, L.
in:
PubMed
|
Google Scholar
|

Published August 1, 2025 - More info
J Clin Invest. 2025;135(15):e191645. https://doi.org/10.1172/JCI191645.
© 2025 Willmann et al. This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
-
Abstract
The immune system must identify genuine threats and avoid reacting to harmless microbes because immune responses, while critical for organismal survival, can cause severe damage and use substantial energy resources. Models for immune response initiation have mostly focused on the direct sensing of microorganisms through pattern recognition receptors. Here, we summarize key features of the leading models of immune response initiation and identify issues they fail to solve individually, including how the immune system distinguishes between pathogens and commensals. We hypothesize and argue that surveillance of disruption to organismal homeostasis and core cellular activities is central to detecting and resolving relevant threats effectively, including infection. We propose that hosts use pattern recognition receptors to identify microorganisms and use sensing of homeostasis disruption to assess the level of threat they pose. We predict that both types of information can be integrated through molecular coincidence detectors (such as inflammasomes or others not yet discovered) and used to determine whether to initiate an immune response, its quality, and its magnitude. This conceptual framework may guide the identification of novel targets and therapeutic strategies to improve the progression and outcome of infection, cancer, autoimmunity, and chronic conditions in which inflammation plays a critical role.
-
Introduction
Infection is one of the most serious and frequent threats to an organism that requires defense mechanisms in the form of immune responses. Regardless of their complexity level, effective immune responses share key mechanistic architectures at the sensing, signal transduction, and effector steps. Sensing is arguably the most conserved and important step across organisms, because it determines whether an immune response should be initiated, the most appropriate type, and its magnitude. The decision to initiate or not is critical because immune responses can harm the host, potentially causing substantial tissue damage or autoimmunity, resulting in organ failure or death (1). Moreover, immune responses are energetically demanding and consume limited resources (2). Therefore, sensing bona fide pathogens while avoiding responses against harmless microorganisms is essential to minimize negative effects on fitness and promote host survival (3). The standard immune response initiation models have mostly focused on directly sensing microorganisms through their molecular motifs via pattern recognition receptors (PRRs). While PRRs have been central to understanding the pathophysiology of immune responses, they sense molecular signatures shared between pathogens and commensal organisms and therefore cannot serve to distinguish them.
Unlike commensals, pathogens disrupt homeostasis in their hosts, often expressed by signs and symptoms of infection. Substantial changes in the set points of strictly controlled physiological parameters are an inevitable consequence of the pathogen’s invasion and life cycle. The information resulting from sensing these changes is likely to integrate with signals that directly identify the presence of a microorganism to shape the decision to initiate an immune response, its quality, and its magnitude. This Review presents a conceptual framework of surveillance immunity that integrates a critical role for mechanisms that sense homeostasis deviations resulting from infection with the initiation and regulation of immune responses. Before we explore this model and discuss its biological and clinical implications, we will briefly describe key features of theoretical models of immune response initiation and identify their main insufficiencies (Figure 1).
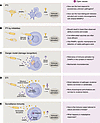 Figure 1
Figure 1Models of innate immune initiation. (A) Pattern-triggered immunity (PTI). Microbial structural molecules (PAMPs or MAMPs) are directly sensed by PRRs, which can activate transcriptional programs or effectors directly. MAMPs that are not conserved or are unknown to the host may not activate PTI. MAMPs may be shared between virulent and avirulent microorganisms (102). (B) PTI by infidelities (14). This model proposes that PRRs are predominantly byproducts of unsuccessful pathogens that lead to biochemical infidelities. This implies a high pressure on pathogens to minimize unsuccessful events and should result in a lower-than-observed ability to evolve and evade (14). Additionally, live-attenuated vaccines tend to have the highest efficiency and sensing of markers of live pathogens (vita-PAMPs) by the host (103). (C) Danger model (damage recognition) (16). PRRs are activated by sensing host molecular patterns released upon compromised tissues. The relevance of DAMPs in the context of infection has not been fully resolved in this model. (D) Effector-triggered immunity (ETI) (21). Virulence factors are sensed by “guard proteins” directly or indirectly by detecting changes or modifications in host proteins (“guardees”). (E) Surveillance immunity (3). Immune responses are triggered by disruption of core cellular functions or homeostasis parameters through stress pathways. Multiple input pathways synergize to generate an output tailored to the nature and level of threat. However, maladaptive responses cannot be fully avoided. Yellow symbols depict microbial factors; purple symbols depict host factors. HAMPs, homeostasis altering molecular processes.
-
Models of immune response initiation
Theoretical models are critical to organizing observations into coherent, explanatory, and predictive frameworks that generate new research hypotheses. Development of these models represents a critical step for new biological discoveries and the potential identification of novel therapeutic approaches informed by knowledge generated from their use.
Pattern-triggered immunity model. The pattern-triggered immunity (PTI) model has been the standard framework for over three decades to explain the mechanisms of immune response initiation (4). From the early formulation of PTI, it follows that germline-encoded PRRs recognize evolutionarily conserved microorganism-associated molecular patterns (MAMPs) (5), motifs associated with molecules that are essential for microorganisms’ survival, but are not produced by the host organism (6). Classic examples are LPS and flagellin, but MAMPs can be functionally and structurally very diverse, ranging from several types of nucleic acids with microbe-specific modifications to proteins, lipids, and carbohydrates (7). PRRs are grouped into families mostly according to their targets and include TLRs, NOD-like receptors (NLRs), RIGI-like receptors (RLRs), other cytosolic nucleic acid sensors, and C-type lectin receptors (CLRs) (reviewed in ref. 7). PRR activation triggers immune signaling pathways that initiate gene expression and metabolic programs, leading to effector responses tailored against each pathogen group, ultimately leading to the generation of long-lasting adaptive immunity (6). These programs are under the control of critical pathways, such as NF-κB, MAPKs, and interferon-regulatory factors (8), that transcribe immune effector molecules like chemokines, cytokines, and interferons and orchestrate an immune and inflammatory response. Other PRRs, like the NLRs, assemble in multimeric complexes, such as the NLR-mediated inflammasome that activates caspases and converts molecular precursors into their bioactive forms.
Experimental observations extensively support this PTI model’s key conceptual components. It has had foundational importance in the field of innate immunity, guiding the mechanistic dissection of its core principles. However, PTI has not resolved central problems in immune response initiation, particularly how the host distinguishes commensals from pathogenic microorganisms. MAMPs are shared by all microorganisms within a specific group, not restricted to pathogens (9, 10). In addition, MAMPs are often described as evolutionarily conserved molecules that microorganisms cannot molecularly change because even small modifications would compromise their viability. However, extensive variations of MAMPs within each group, or even species, are frequent and represent a strategy for pathogens to avoid or antagonize detection (9, 11). The recent demonstration that organisms fail to respond to pathogens with known MAMPs but no common evolutionary history further supports this argument (12). The barcode hypothesis proposes that microorganisms of high pathogenic potential could be recognized by their distinct combinations of MAMP patterns, which would allow for tailor-made responses (13). However, such combinations cannot categorize most pathogens, suggesting additional cues for pathogen distinction are necessary (9).
Therefore, the PTI model is insufficient to explain how hosts distinguish between infection and colonization, viable and dead, and pathogenic and nonpathogenic microorganisms. It also does not provide mechanistic insights into how the host assigns the quality, magnitude, and duration of an immune response in the face of a perceived infectious threat. Therefore, PRR activation is not sufficient. Additional signals are required to shape an effective immune response and to minimize collateral tissue damage.
Infidelities model. According to the infidelities model, PRRs detect pathogens via MAMPs, often resulting from microbial biochemical infidelities or mistakes during the infectious life cycle (14). Incomplete or erroneous microbial processes, like the release of incomplete viral genomes or misdirected bacterial components, still activate immune responses. This model proposes that PRRs may not target pathogens directly but instead detect the products of such errors (14). Although decreasing these errors would minimize detection, some level of error is critical for microbial evolvability (and, consequently, survival), so pathogens cannot eliminate or even excessively lower their rate (14). The model posits that exceptions, including dedicated TLR detection of functional LPS, are relatively recent evolutionary events. If correct, the infidelities model’s predictions may guide the development of more effective immunotherapies and antibiotic drugs. Novel strategies could be based on superior modes of PRR activation or on targeting pathogen infidelities. However, this model is not supported by the observation that live-attenuated vaccines are more effective at generating protective immune responses than those that use inactivated or subcomponents of the pathogen. These observations instead support the idea that sensing indicators of pathogen viability and infectivity synergize with MAMP sensing to elicit effective and vigorous immune responses (discussed below).
Patterns of pathogenesis. The patterns of pathogenesis (9) concept proposes that the immune system responds to MAMPs by contextualizing additional signals. Directly sensing microorganisms alone is insufficient for distinguishing pathogens from commensal microorganisms and selecting an appropriate immune response. Additional signals may derive from factors that pathogens use to infect their hosts, multiply, and later spread to additional hosts. Sensing additional microorganism characteristics and the consequences of their presence may help hosts form an assessment of pathogen virulence (Figure 2), influencing immune response initiation and calibration based on the threat level. For example, virulence factors like pore-forming toxins and bacterial secretion systems may strongly signal pathogenicity and activate an immune response (reviewed in ref. 15). It has been proposed that the threat level to the host and the necessary immune response quality and magnitude can be assessed by integrating at least five checkpoints (Figure 2, reviewed in ref. 7). These include the integration of tissue-specific signals and the distinction between (a) soluble and particulate MAMPs, (b) viable and dead microorganisms, (c) appropriate spatial location of microorganisms, (d) invasive and noninvasive microorganisms, and (e) pathogenic and nonpathogenic microorganisms. Within the framework of patterns of pathogenesis, the danger, the effector-triggered immunity, and the surveillance immunity models describe possible paradigms of immune response initiation.
 Figure 2
Figure 2Patterns of pathogenesis. The risk level (threat to host system homeostasis) and the magnitude and nature of the immune response that needs to be activated are assessed using direct sensing of microorganisms and additional contextual signals (9). The pathogen must overcome several checkpoints (depicted in columns labeled Checkpoint 1–5) before it poses the highest level of threat, resulting in a vigorous immune response (7). Checkpoint 1: Soluble MAMPs initiate cytokine and chemokine production remotely, while MAMPs on whole microorganisms trigger direct microbicidal responses. Checkpoint 2: Vita-PAMPs, such as bacterial mRNA, indicate live microorganisms capable of growth, multiplication, and invasion and trigger enhanced immune responses by activating PRRs. Checkpoint 3: The need and type of immune response to microbial presence varies according to the tissue’s physiology and microenvironment, ensuring appropriate responses. Systemic threats trigger immediate, strong reactions to prevent severe consequences, while local tissue responses are tightly regulated. At the subcellular level, the strongest immune responses are initiated against agents that invade the cytosol. Checkpoint 4: The degree of invasiveness is critical information for the immune system to distinguish between pathogenic and nonpathogenic microorganisms. While commensal bacteria coexist with the host without causing disease, they can become pathogenic if they breach sterile tissues. Invasive forms of microbes expose specific molecules or morphologies that signal potential threats, leading to more robust immune activation. Commensals can act as facultative pathogens under specific conditions. Commensal bacteria can become invasive due to host factors like immunodeficiency, pregnancy, or treatments altering the microenvironment. The immune system and intact physical barriers are crucial for preventing this switch. Invasiveness can be controlled by inhibiting the quorum-sensing system of microorganisms. Checkpoint 5: Virulence. Microorganisms are classified as pathogens or nonpathogens based on their ability to cause disease, correlating with virulence factors that disrupt host barriers and invade tissues.
Danger model. The danger model (16) states that the immune system recognizes pathogens through the consequences of their presence by identifying damage-associated molecular patterns (DAMPs), endogenous molecules released from host cells due to cell death or damage (reviewed in ref. 17). The immune system would recognize the damage caused by pathogens, not the microorganisms that cause it (18). This model has been useful in the context of sterile inflammation but is insufficient in infection, where mechanistic inconsistencies, especially its initiation step, remain unresolved (18). It will be interesting to investigate mechanisms through which the host may be able to distinguish between sterile injury and injury caused by pathogens or the immune response to eliminate them.
Effector-triggered immunity model. The effector-triggered immunity (ETI) model was initially defined in plants as a protective type of immune response against microbial effectors (19). Because PRR signaling alone provides insufficient information about microorganismal threat level, sensing virulence factors is a critical component of immune response initiation (Figure 2) (15, 20). Detection of pathogen-encoded virulence factors most often occurs indirectly through the sensing of their virulence activities (reviewed in ref. 21). Examples are (a) the guard hypothesis (22), wherein a virulence factor can modify a target protein (the “guardee”) that is identified by a sensor (the “guard”); (b) a virulence factor is directly identified by a host sensor (an example mostly restricted to the case of plants); (c) a virulence factor causes cellular stress; and (d) the pathogen activity perturbs or eliminates a protein that is an inhibitor of immune responses. ETI was reviewed in ref. 21 and, therefore, will not be analyzed in detail here. Although the scope of ETI is expanding (15, 20), creating considerable conceptual overlap between ETI and surveillance immunity, here we take the stricter definition of ETI, proposing that virulence factors from pathogens are the key features used for their recognition (15, 20), distinguished from surveillance immunity pathways initiated by a broader range of stimuli, even those beyond infection (discussed below).
Overall, the models summarized above do not resolve key questions relating to immune response initiation. In recent years, several other theoretical models that integrate physiology principles have been proposed to address these issues and attempt a unifying and coherent framework where immunity is a central component of the many physiological processes that maintain homeostasis. These include a framework for homeostasis maintenance (23), the discontinuity theory of immunity (24, 25), the equilibrium model (26–28) and the quantal theory of immunity (29). We (3) and others (30, 31) have previously proposed a model of surveillance immunity for the initiation of innate immune responses and homeostasis maintenance. In the following sections, we describe, update, and discuss the biological and clinical implications of this model (Figure 3) that focuses on the central role of physiological disruptions for the initiation and quality of immune responses.
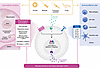 Figure 3
Figure 3Surveillance immunity. All major groups of pathogens (viruses, bacteria, protozoan parasites, and fungi) trigger stress responses to core homeostatic processes such as DNA damage, replicative stress, mitochondrial dysfunction and proteostatic stress (UPRmt), translation inhibition and ER proteostatic stress (UPRer), in addition to direct recognition by PRRs. Direct and indirect sensing of homeostasis disruption and signaling by PRR is integrated and synergizes in the production of immune and homeostasis effectors to tailor effector responses to specific classes of pathogens and level of threat. Both disease resistance (directed against the pathogen) and disease tolerance mechanisms that act on the host (to limit tissue damage, collateral damage, and tissue dysfunction) are activated. ER, endoplasmic reticulum.
-
Surveillance immunity model hypothesis
Homeostasis is a dynamic and self-regulating process that allows organisms to actively maintain a stable internal environment in response to changing internal and external conditions, using negative feedback mechanisms (3, 32). Survival and functional preservation require keeping several physiological parameters within strictly enforced ranges (3, 32). First-line homeostatic circuits are disrupted in response to large internal or external perturbations, such as severe systemic infection. In these cases, negative feedback mechanisms are insufficient to maintain key regulated variables within the required ranges (33) and need instead feed-forward mechanisms like inflammation that coordinate emergency responses to restore homeostasis (34) but may activate a hyperinflammatory state in the host as a result.
Surveillance immunity model. Our proposed surveillance immunity model hypothesizes that organisms integrate the information coming from the direct sensing of a microorganism, mostly via its molecular signatures using PRRs, with information about physiologic disruption resulting from the activities of the pathogen. This enables the host to assess the level of threat and gauge the need to initiate an immune response, its quality, and magnitude. We predict that while initial physiological perturbations will be caused by the pathogen alone, once the immune response is initiated, immune-driven physiological disruptions (including those caused by collateral tissue damage or cytokine production) will also contribute to a possible feed-forward process to shape and amplify the immune response or to terminate it. For example, some cytokines will induce fever, which is known to promote pathogen control by many processes, including increasing T cell proliferation, cell migration, and antigen presentation and restricting pathogen replication (35).
In the extreme, organisms may initiate inflammation and full-blown immune responses owing to substantial deviations in homeostasis alone (30). This framework implies that initiating an immune response requires contextual information that accompanies the presence of a microorganism. The contribution of additional signals, or even their self-sufficiency for immune response initiation, is particularly well illustrated by the fact that organisms that lack bona fide PRRs, like Caenorhabditis elegans, initiate aversive behaviors to pathogens and are capable of mounting effective immune and detoxification responses against them (30). Multiple pathways for detoxification, pathogen response, and mitochondrial repair were first discovered in C. elegans, including ceramide biosynthesis and the mevalonate pathways (36), and nuclear hormone receptor-dependent detoxification genes (37). Interestingly, mitochondrial dysfunction triggers RNA interference in C. elegans through a pathway homologous to the mammalian RIG-I antiviral response (38).
The initiation of immune responses based on the information provided by sensors of substantial physiological disruption may be able to detect a wide range of relevant threats, regardless of the initiating molecular signatures, using a small set of genome-encoded components (3). In addition to providing a critical contribution to the initiation step of an immune response, it is likely that early sensing of substantial deviations in homeostasis parameters can also play a critical role in limiting tissue damage and the activation of tissue damage repair to preserve organ function and to allow the return to steady state (39).
We propose that coincidence detectors may mechanistically mediate the integration of signals from PRRs and sensors of homeostasis disruption (Figure 3). The inflammasome is currently the best example to function as a coincidence detector. The effector function of inflammasomes requires two signals. The first leads to NF-κB activation and may be initiated by directly sensing microorganisms using PRRs. The second activating signal is often given by potassium efflux across the plasma membrane resulting from the effect of microbial toxins that disrupt ion gradients across the membrane, as in the case of Staphylococcus aureus α-toxin (40), which can indicate membrane disruption by pathogen activity. This type of mechanism applies not only to the plasma membrane, but also to intracellular membranes; for instance, the influenza virus M2 protein is a proton-selective ion channel that neutralizes the pH of the trans-Golgi network (41). Other good candidate molecules for coincidence detectors, including those with a known role in immune responses, are the TIR-domain-containing adaptor protein-inducing IFN-β (TRIF), as well as the PRRs NOD1/2, which sense bacterial peptidoglycans (42). In addition to their role as PRRs, NOD1/2 also act as metabolic sensors of stress by responding to the endogenous metabolite sphingosine-1-phosphate (S1P) that is produced in response to cellular perturbations (43). Notably, while S1P binds directly to the nucleotide-binding domains of NOD1/2 to activate NF-κB signaling, peptidoglycan sensing is achieved through the leucine-rich repeats domain of NODs (43), meaning that a PRR molecule can act as a coincidence detector integrating bacterial and metabolic cues for optimal activation of downstream inflammatory responses. TRIF can also act as a coincidence detector by linking the TLR and NLRP3 inflammasome pathways using a different mechanism: TRIF is critical for production of high concentrations of IFN-β in response to the mRNA vita-PAMP, which distinguishes dead from live bacteria and informs the host organism on the threat posed by the presence of a microorganism (44). One of the main downstream effector roles of coincidence detectors like TRIF and inflammasomes may be the coupling of transcriptional and posttranslational processes, possibly initiated by different signals resulting from sensing of MAMPs and physiologic disruption, which are required to produce most immune effectors.
Another key prediction of the surveillance immunity model is that pathogens, unlike commensals, cause substantial metabolic stress in the host. Tightly regulated self-metabolites, which can be sensed directly or indirectly, undergo large deviations from the homeostatic set points. For instance, as described above, NOD1/2 sensing of S1P results from cellular stress that increases in response to disruption of cellular homeostasis by the presence of a pathogen (43). In addition to sensing self-metabolites, the host can directly sense a repertoire of non-self-metabolites produced by microorganisms, which may serve as a measure of their pathogenic potential and, therefore, distinguish commensals from pathogenic microorganisms. For example, C. elegans uses the NHR-86 nuclear hormone receptor (a homolog of mammalian hepatocyte nuclear factor 4 [HNF4], which has roles in glucose and lipid metabolism in insects, ref. 45) to sense the non-self, toxic pathogen–derived phenazine-1-carboxamide metabolite produced by pathogenic strains of Pseudomonas aeruginosa and activate innate immune responses (46). In addition, C. elegans can initiate a behavioral avoidance response of pathogens following the olfactory sensing of volatile compounds from pathogens like P. aeruginosa (47). An avoidance response to E. faecalis in C. elegans can also result from activating TRPM channels that mediate learned pathogen avoidance that causes intestinal distention (48).
Notably, hepatic HNF4α has now been implicated in polymicrobial sepsis-associated metabolic reprogramming, where it is required to prevent liver steatosis and organ damage while inducing liver regeneration, thereby decreasing the risk of death (49). Phenazines, a group of bacterial virulence factors, were identified as ligands for the aryl hydrocarbon receptor, which recognizes a wide array of endogenous ligands and environmental toxins and initiates immune responses in mammals (50). Intestinal tuft cells utilize taste and other metabolite receptors (including for succinate) that enables them to act as mucosal sentinel cells and activate type 2 immune responses (51). Other GPCRs have also been implicated in sensing self-metabolites (reviewed in ref. 2), in some cases participating in inflammasome activation, e.g., OLFR2 (52), in a manner that is compatible with the concept of coincidence detection. Interestingly, the NLRP3 inflammasome can respond to the microbial danger signals butyrate and propionate (53). Microbiota-derived metabolites, like butyrate, can modulate intestinal (type 2) immunity, for example, by restricting tuft cell differentiation (54).
Immune responses can be initiated locally by cell-autonomous perturbation of core cellular functions or in distant tissues from the initial site of homeostasis disruption, implicating non-cell-autonomous stress responses and interorgan communication in the mechanisms of surveillance immunity and promoting survival to environmental challenges that threaten the integrity of their genome, proteome, or metabolome (55). The stressed tissue may secrete factors that transmit signals to tissues in different organs and initiate processes that help the organism cope with stress. For example, evidence in C. elegans shows that the mitochondrial unfolded protein response (UPRmt) can be non-cell-autonomously mediated by Wnt signaling, which relays mitochondrial stress signals (“mitokines”) from neurons to peripheral tissues (56). Mild muscle mitochondrial distress in D. melanogaster initiates both local (redox-dependent induction of genes that regulate the UPRmt) and systemic responses (involving the transcriptional induction of the Drosophila ortholog of insulin-like growth factor–binding protein 7) that antagonize insulin signaling and promote mitophagy (57). IL-6 is induced in response to several forms of physiological disruption, including those caused by infection to coordinate systemic immunometabolic reprogramming; it behaves as a systemic stress hormone that mediates interorgan axis, such as those between brain/brown fat/liver (58). These and other types of responses cooperate to induce immune responses, cytoprotective pathways, and repair responses critical to dealing with stressors, ranging from environmental toxins to infectious challenges (59), that limit tissue damage and ultimately prolong lifespan.
We next explore examples of surveillance immunity in response to diverse types of homeostasis disruption at multiple organismal levels.
-
Disruption of systemic and metabolic homeostasis
Arterial partial pressures of O2 and CO2, concentrations of K+, Ca2+, H+ (pH) and blood glucose, core body temperature, mean arterial pressure, blood volume, and blood osmolality are critical homeostatic variables. The organism monitors these variables and counters deviation using negative feedback mechanisms (33). Substantial deviations of these parameters have been documented to lead to inflammatory responses, as negative feedback mechanisms are insufficient to bring them back to their original physiological ranges.
Low O2 (hypoxia) and glucose concentrations. Low O2 (hypoxia) and glucose concentrations (which occur in pathological niches like tumors and infected or ischemic tissues, ref. 60) decrease the function of the rate-limiting enzyme of the mevalonate kinase pathway (HMG-CoA reductase, HMGCR), which leads to the activation of NLRP3 inflammasome (61). Stroke induces sustained inflammation and drives atherosclerosis by activating Notch1 in endothelial cells (62). Prolonged hypoxia can be sensed by pyridoxine 5′-phosphate oxidase (PNPO), an enzyme that catalyzes the bioactivation of vitamin B6, which decreases its activity under prolonged hypoxia, leading to deficient lysosome acidification and delayed resolution of the inflammatory response (63). Hypoxia is also a known cause of pulmonary hypertension because macrophages activate vascular remodeling (64).
Hemodynamic perturbations. Hemodynamic perturbations, like blood pressure increase, can cause microglial inflammatory activation, which can then act to control blood pressure changes (65, 66). The mechanically activated ion channels PIEZO1 and PIEZO2 are the critical sensors in baroreceptor neurons that monitor blood pressure to keep it in the appropriate physiological range (67). PIEZO1 is present and has demonstrated roles in macrophage-initiated inflammatory responses (68, 69). Notably, Piezo1 deletion in the myeloid compartment decreases macrophage kidney infiltration and activation to prevent renal fibrosis, a common consequence of chronic hypertension (70).
Thermoregulation. Pathogens often disrupt thermoregulation by inducing either fever or hypothermia. Fever-range heat constitutes a danger signal that causes mitochondrial stress, resulting in the increase of T cell proliferation, migration, and inflammatory functions (35).
Metabolic reprogramming. Metabolic reprogramming in the presence of a pathogen may provide key signals to initiate and regulate immune responses (2). Sensing of substantial deviations in controlled metabolic fluxes may signal the presence of specific pathogen groups, because each has requirements that vary according to the specificities of their life cycles. Notably, blood glucose concentrations are modulated by infection. The host can sense these changes and potentiate innate antiviral immune responses (71) as well as metabolic defense strategies (72). A shift toward glycolytic-based metabolism is a hallmark of resistance mechanisms, while fatty acid oxidation and oxidative phosphorylation are central for disease tolerance mechanisms (2). Interestingly, acute suppression of mitochondrial ATP production prevents apoptosis and provides an essential signal of NLRP3 inflammasome activation (73), which may constitute an example of coincidence detection.
Amino acid. Amino acid availability can be sensed and interpreted by the host as the presence of a pathogen and constitutes a central regulatory node for immune responses and infection pathophysiology to bacteria and viruses. Host sensing of amino acid depletion induced by invasive bacterial pathogens initiates protective innate immune and stress responses, including by a decrease in mTOR activity leading to autophagy (74). Virus-dependent activation of GCN2, a conserved serine/threonine kinase that works as a stress sensor in response to amino acid deficiency, initiates autophagy and enhances antigen presentation to CD4+ and CD8+ T cells (75). Serine metabolism is critical in antiviral immunity and constitutes an integration hub for cellular metabolism, antiviral immunity, and epigenetic regulation. Deficiency of the amino acid serine promotes virus-induced IFN-β production (76). By contrast, increases in serine suppress interferon responses (77). Similarly, methionine restriction has been observed to limit tumor growth and to increase the responses to anticancer therapies (78).
Nucleotide depletion. Nucleotide depletion promotes cell fate transitions and induces DNA replication stress (79). In C. elegans, perturbations in purine metabolism are sensed and act as signals to promote defense against epithelial infection (80). Similarly, cellular pyrimidine deficiency triggers mitochondrial DNA–dependent innate immunity (81). NAD+ depletion is sensed by the innate immune sensor NLRC5 to trigger PANoptosis (a caspase and RIPK-driven inflammatory cell death mediated by PANoptosomes) and inflammation (82).
Both a decrease in cholesterol synthesis (83) and excess cholesterol concentrations (84, 85) have been causally linked to enhanced immune responses, suggesting that substantial deviations in cholesterol concentrations trigger inflammatory responses, possibly reflecting the targeting of cholesterol synthesis pathways by pathogens, especially viruses. Several mechanisms are likely to prevent deviations in cholesterol concentrations. Genetic (86) or pharmacologic (83) cholesterol synthesis inhibition by statins greatly increases interferon responses. By contrast, cholesterol excess directly causes mitochondrial DNA release and consequent activation of the AIM2 inflammasome, which can be prevented by producing 25-hydroxycholesterol in activated macrophages (84). Excess cholesterol has also been shown to promote adipose tissue inflammation (87). Oxidized lipids (OLs) may serve as generic indicators of threat to the host, both in the context of sterile and septic injury. OLs resulting from tissue injury caused by infection can act as immunomodulatory signals, leading to pro- or antiinflammatory downstream responses, depending on the context (88). Similarly, in Drosophila, sugar alcohols of the polyol pathway may serve as alarmins and mediate communication between local and systemic innate immune responses (89).
-
Disruption of cellular, organellar and molecular homeostasis
We have previously documented evidence supporting a critical role for the disruption of homeostasis at the cellular, organellar, and molecular levels in shaping the initiation and quality of immune responses (reviewed in ref. 3). In Table 1, we summarize, update, and document the clinical implications of these observations. Key cellular processes, such as energy metabolism and protein production, are compartmentalized within organelles. Damage to organelles can cause leakage of contents that activate sensors that detect misplaced molecules. Such disruptions can result from infections or sterile conditions, like autoinflammatory diseases. Stresses like infections, mechanical strain, or nutrient changes disrupt organelle integrity, triggering repair programs to restore balance. Persistent damage often leads to low-grade inflammation (parainflammation), while transient stress mechanisms that promote inflammation remain less understood (34). Emerging evidence highlights mitochondria, ER, lysosome, Golgi, and nuclear envelope stress as sources of proinflammatory signals (Table 1). Targeting pathways that restore cellular, organellar, and molecular homeostasis or mitigate stress responses offers therapeutic potential for infections and chronic inflammatory diseases.
-
Conclusions and future perspectives
Initiation of immune responses following the distinction between pathogens and commensal microorganisms is likely to require direct sensing of microorganisms in the context of the physiological perturbations they cause. Integration of this information may be accomplished at the molecular level by coincidence detectors that signal for downstream events only when both types of signals are present. There are many open questions posed by a model of surveillance immunity (Table 2), including (a) the identity of upstream sensors of pathogen-disrupted physiological parameters; (b) how these signals molecularly integrate with the information coming from direct microorganism sensing; (c) the identity of coincidence detectors; and (d) the downstream signaling events leading to resistance and disease tolerance processes. This knowledge is not only of biological interest but has important clinical implications, including the distinction between colonization and infection. It can also potentially inspire novel therapeutic strategies to treat severe infection and chronic inflammation.
Inflammation is a core component of most, if not all, known pathologic chronic conditions (39). It has historically been considered a response to septic and aseptic injury, but it may have evolved as an adaptive response for restoring homeostasis, as it is now clear that inflammation is likely to have additional critical physiological roles, including in the orchestration of the feed-forward mechanisms that deal with severe disruption in homeostasis that cannot be restored by negative feedback mechanisms (34). Macrophages are likely to have central roles in sensing physiological disruption that initiates inflammatory responses (90).
If, as we argue here, sensing of homeostasis disruption caused by the presence of a pathogen is critical, perhaps sufficient, to initiate immune responses, it is conceivable that the persistence of physiologically monitored parameters outside of their normal ranges may cause systemic chronic inflammation that favors the occurrence of the highly prevalent modern chronic conditions, including type 2 diabetes and cardiovascular diseases. In this context, for example, maintaining high blood pressure or nontreated sleep apnea, which causes repeated decreases in O2 saturation, would promote the initiation and progression of cardiovascular disease mainly because they cause persistent systemic inflammation. Other forms of persistent nonresolved nutritional and metabolic stress (91) could lead to many of the other current major health concerns, including obesity and metabolic dysfunction–associated steatohepatitis (92). In addition, chronic inflammation can be a critical contributing factor to many cancer types. Notably, inflammation is not routinely treated in these conditions, but several observations, including in human clinical trials, suggest that targeting inflammation may be highly effective in preventing acute cardiovascular events, including myocardial infarction (93) and stroke (94).
An additional important clinical implication of a model of surveillance immunity is the need for research to understand the effects of many drugs used routinely by hundreds of millions of people, either to explore their nonconventional effects or to minimize their undesirable side effects. For example, clinicians have known and empirically preferentially used classes of antibiotics that better resolve an infection than would be expected from their direct antimicrobial activity alone and are superior to different classes with overlapping antimicrobial spectra. In addition to their direct antibacterial activities, these antibiotics have been demonstrated to have off-target host effects that cause physiologic perturbations, including mitochondrial protein synthesis inhibition and DNA damage that trigger the production of immune mediators (often interferon-stimulated genes) or limit tissue damage caused by the infection or associated immune response, thereby promoting organ function (reviewed in ref. 2). Notably, antibiotics, like tetracyclines, are among the most prescribed drugs for dermatological conditions and act via mechanisms independent of their antibiotic activity, which likely rely on their ability to cause physiologic perturbations. Statins, which block cholesterol synthesis, modulate immune responses and, therefore, may impact cardiovascular diseases beyond their well-known direct cholesterol-lowering properties. Another example is that of highly prescribed drugs that affect the function of the mitochondrial ETC, like metformin. ETC perturbations have been shown to modify the progression of a severe infection (95). These properties should be further explored in relation to effects on infection susceptibility and vaccine effectiveness. We can also expect that cancer chemotherapeutic drugs that affect nucleic acid homeostasis, particularly DNA and chromatin, and cell death pathways with DAMP release, dramatically affect immune (96) and inflammatory responses (97), which may be explored to get more effective antitumoral responses and decrease resistance to standard therapy (98), in addition to the management of undesired side effects (99).
Models of immune response initiation, particularly those relying solely on the direct detection of microorganisms, inadequately describe the pathophysiological mechanisms governing the initiation, progression, and resolution of immune responses. Models that consider homeostasis disruption as a key factor in immune response initiation are necessary not only because they add considerably to our understanding of organismal physiology, but also because they have broad implications for human pathophysiology and effective treatment of acute infection and chronic disease.
-
Acknowledgments
KW is supported by an Fundação para a Ciência e a Tecnologia (FCT) CEEC individual contract (CEECIND/02661/2018) and by an FCT exploratory grant (EXPL/MED-OUT/0745/2021). LFM is supported by an FCT CEEC individual contract (CEECIND/03812/2017). Work in the Ferreira Moita laboratory is supported by grants from the Oeiras-ERC Frontier Research Incentive Award and ‘‘la Caixa’’ Foundation (LCF/PR/HR23/52430007).
Address correspondence to: Luis F. Moita, Faculdade de Medicina, Universidade de Lisboa, School of Medicine, University of Lisbon, Avenida Professor Egas Moniz, 1649-028 Lisboa, Portugal. Email: lmoita@medicina.ulisboa.pt.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2025, Willmann et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2025;135(15):e191645. https://doi.org/10.1172/JCI191645.
-
References
- Soares MP, et al. Disease tolerance and immunity in host protection against infection. Nat Rev Immunol. 2017;17(2):83–96.
- Willmann K, Moita LF. Physiologic disruption and metabolic reprogramming in infection and sepsis. Cell Metab. 2024;36(5):927–946.
- Colaço HG, Moita LF. Initiation of innate immune responses by surveillance of homeostasis perturbations. FEBS J. 2016;283(13):2448–2457.
- Janeway CA. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(1):1–13.
- Medzhitov R. Approaching the asymptote: 20 years later. Immunity. 2009;30(6):766–775.
- Medzhitov R, Janeway CA Jr. Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91(3):295–298.
- Blander JM, Sander LE. Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat Rev Immunol. 2012;12(3):215–225.
- Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454(7203):428–435.
- Vance RE, et al. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6(1):10–21.
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449(7164):819–826.
- Jain S, Darveau RP. Contribution of Porphyromonas gingivalis lipopolysaccharide to periodontitis. Periodontol 2000. 2010;54(1):53–70.
- Gauthier AE, et al. Deep-sea microbes as tools to refine the rules of innate immune pattern recognition. Sci Immunol. 2021;6(57):eabe0531.
- Aderem A. Phagocytosis and the inflammatory response. J Infect Dis. 2003;187 Suppl 2(s2):340–345.
- Kagan JC. Infection infidelities drive innate immunity. Science. 2023;379(6630):333–335.
- Lopes Fischer N, et al. Effector-triggered immunity and pathogen sensing in metazoans. Nat Microbiol. 2020;5(1):14–26.
- Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045.
- Huang Y, et al. DAMP sensing and sterile inflammation: intracellular, intercellular and inter-organ pathways. Nat Rev Immunol. 2024;24(10):703–719.
- Józefowski S. The danger model: questioning an unconvincing theory. Immunol Cell Biol. 2016;94(2):164–168.
- Jones J. The plant immune system. Nature. 2006;444(7117):323–329.
- Kufer TA, et al. Guardians of the cell: effector-triggered immunity steers mammalian immune defense. Trends Immunol. 2019;40(10):939–951.
- Remick BC, et al. Effector-triggered immunity. Annu Rev Immunol. 2023;41:453–481.
- Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411(6839):826–833.
- Germain RN. Maintaining system homeostasis: the third law of Newtonian immunology. Nat Immunol. 2012;13(10):902–906.
- Pradeu T, Vivier E. The discontinuity theory of immunity. Sci Immunol. 2016;1(1):AAG0479.
- Pradeu T, et al. The speed of change: towards a discontinuity theory of immunity? Nat Rev Immunol. 2013;13(10):764–769.
- Eberl G, Pradeu T. Towards a general theory of immunity? Trends Immunol. 2018;39(4):261–263.
- Eberl G. Immunity by equilibrium. Nat Rev Immunol. 2016;16(8):524–532.
- Eberl G. A new vision of immunity: homeostasis of the superorganism. Mucosal Immunol. 2010;3(5):450–460.
- Smith KA. The quantal theory of immunity. Cell Res. 2006;16(1):11–19.
- Melo JA, Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell. 2012;149(2):452–466.
- Pukkila-Worley R. Surveillance immunity: an emerging paradigm of innate defense activation in Caenorhabditis elegans. PLoS Pathog. 2016;12(9):e1005795.
- Modell H, et al. A physiologist’s view of homeostasis. Adv Physiol Educ. 2015;39(4):259–266.
- Kotas ME, Medzhitov R. Homeostasis, inflammation, and disease susceptibility. Cell. 2015;160(5):816–827.
- Chovatiya R, Medzhitov R. Stress, inflammation, and defense of homeostasis. Mol Cell. 2014;54(2):281–288.
- Wilander BA, Rathmell JC. Metabolic and stress response adaptations in T cells to fever and physiological heat. Trends Immunol. 2025;46(3):195–205.
- Liu Y, et al. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 2014;508(7496):406–410.
- Mao K, et al. Mitochondrial dysfunction in C. Elegans activates mitochondrial relocalization and nuclear hormone receptor-dependent detoxification genes. Cell Metab. 2019;29(5):1182–1191.
- Mao K, et al. Mitochondrial dysfunction induces RNA interference in C. elegans through a pathway homologous to the mammalian RIG-I antiviral response. PLoS Biol. 2020;18(12):e3000996.
- Medzhitov R. The spectrum of inflammatory responses. Science. 2021;374(6571):1070–1075.
- Muñoz-Planillo R, et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38(6):1142–1153.
- Ichinohe T, et al. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11(5):404–410.
- Inohara N, et al. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74(1):355–383.
- Pei G, et al. Cellular stress promotes NOD1/2-dependent inflammation via the endogenous metabolite sphingosine-1-phosphate. EMBO J. 2021;40(13):e106272.
- Sander LE, et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474(7351):385–389.
- Wang K, et al. A hepatocyte nuclear factor BtabHNF4 mediates desiccation tolerance and fecundity in whitefly (Bemisia tabaci). Environ Entomol. 2023;52(1):138–147.
- Peterson ND, et al. Non-canonical pattern recognition of a pathogen-derived metabolite by a nuclear hormone receptor identifies virulent bacteria in C. elegans. Immunity. 2023;56(4):768–782.
- Prakash D, et al. 1-Undecene from Pseudomonas aeruginosa is an olfactory signal for flight-or-fight response in Caenorhabditis elegans. EMBO J. 2021;40(13):e106938.
- Filipowicz A, et al. TRPM channels mediate learned pathogen avoidance following intestinal distention. Elife. 2021;10:e65935.
- Van Dender C, et al. A critical role for HNF4α in polymicrobial sepsis-associated metabolic reprogramming and death. EMBO Mol Med. 2024;16(10):2485–2515.
- Moura-Alves P, et al. AhR sensing of bacterial pigments regulates antibacterial defence. Nature. 2014;512(7515):387–392.
- Schneider C, et al. Regulation of immune responses by tuft cells. Nat Rev Immunol. 2019;3(1):584–593.
- Orecchioni M, et al. Olfactory receptor 2 in vascular macrophages drives atherosclerosis by NLRP3-dependent IL-1 production. Science. 2022;375(6577):214–221.
- Wang W, et al. Butyrate and propionate are microbial danger signals that activate the NLRP3 inflammasome in human macrophages upon TLR stimulation. Cell Rep. 2024;43(9):114736.
- Eshleman EM, et al. Microbiota-derived butyrate restricts tuft cell differentiation via histone deacetylase 3 to modulate intestinal type 2 immunity. Immunity. 2024;57(2):319–332.
- Owusu-Ansah E, Perrimon N. Stress signaling between organs in metazoa. Annu Rev Cell Dev Biol. 2015;31(1):497–522.
- Zhang Q, et al. The mitochondrial unfolded protein response is mediated cell-non-autonomously by retromer-dependent Wnt signaling. Cell. 2018;174(4):870–883.
- Owusu-Ansah E, et al. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155(3):699–712.
- Qing H, et al. Origin and function of stress-induced IL-6 in murine models. Cell. 2020;182(2):372–387.
- Shore DE, et al. Induction of cytoprotective pathways is central to the extension of lifespan conferred by multiple longevity pathways. PLoS Genet. 2012;8(7):e1002792.
- Taylor CT, Colgan SP. Regulation of immunity and inflammation by hypoxia in immunological niches. Nat Rev Immunol. 2017;17(12):774–785.
- Raulien N, et al. Glucose-oxygen deprivation constrains HMGCR function and Rac1 prenylation and activates the NLRP3 inflammasome in human monocytes. Sci Signal. 2024;17(845):add8913.
- Liu M, et al. Brain ischemia causes systemic Notch1 activity in endothelial cells to drive atherosclerosis. Immunity. 2024;57(9):2157–2172.
- Sekine H, et al. PNPO-PLP axis senses prolonged hypoxia in macrophages by regulating lysosomal activity. Nat Metab. 2024;6(6):1108–1127.
- Kumar R, et al. Monocytes and interstitial macrophages contribute to hypoxic pulmonary hypertension. J Clin Invest. 2025;135(6):e176865.
- Rao Y, Peng B. Microglia bridge brain activity and blood pressure. Immunity. 2024;57(9):2000–2002.
- Wei B, et al. Microglia in the hypothalamic paraventricular nucleus sense hemodynamic disturbance and promote sympathetic excitation in hypertension. Immunity. 2024;57(9):2030–2042.
- Zeng WZ, et al. PIEZOs mediate neuronal sensing of blood pressure and the baroreceptor reflex. Science. 2018;362(6413):464–467.
- Tang Y, et al. Mechanosensitive Piezo1 protein as a novel regulator in macrophages and macrophage-mediated inflammatory diseases. Front Immunol. 2023;14:1149336.
- Xie Y, Hang L. Mechanical gated ion channel Piezo1: Function, and role in macrophage inflammatory response. Innate Immun. 2024;30(2-4):32–39.
- He Y, et al. Myeloid Piezo1 deletion protects renal fibrosis by restraining macrophage infiltration and activation. Hypertension. 2022;79(5):918–931.
- Šestan M, et al. An IFNγ-dependent immune-endocrine circuit lowers blood glucose to potentiate the innate antiviral immune response. Nat Immunol. 2024;25(6):981–993.
- Weis S, et al. Metabolic adaptation establishes disease tolerance to sepsis. Cell. 2017;169(7):1263–1275.
- Saller BS, et al. Acute suppression of mitochondrial ATP production prevents apoptosis and provides an essential signal for NLRP3 inflammasome activation. Immunity. 2025;58(1):90–107.
- Tattoli I, et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 2012;11(6):563–575.
- Ravindran R, et al. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science. 2014;343(6168):313–317.
- Shen L, et al. Serine metabolism antagonizes antiviral innate immunity by preventing ATP6V0d2-mediated YAP lysosomal degradation. Cell Metab. 2021;33(5):971–987.
- Agerer B, et al. The serine’s call: Suppressing interferon responses. Cell Metab. 2021;33(5):849–850.
- Ji M, et al. Dietary methionine restriction in cancer development and antitumor immunity. Trends Endocrinol Metab. 2024;35(5):400–412.
- Do BT, et al. Nucleotide depletion promotes cell fate transitions by inducing DNA replication stress. Dev Cell. 2024;59(16):2203–2221.
- Tecle E, et al. The purine nucleoside phosphorylase pnp-1 regulates epithelial cell resistance to infection in C. elegans. PLoS Pathog. 2021;17(4):e1009350.
- Sprenger HG, et al. Cellular pyrimidine imbalance triggers mitochondrial DNA-dependent innate immunity. Nat Metab. 2021;3(5):636–650.
- Sundaram B, et al. NLRC5 senses NAD+ depletion, forming a PANoptosome and driving PANoptosis and inflammation. Cell. 2024;187(15):4061–4077.
- Nishimura T, et al. Cholesterol restriction primes antiviral innate immunity via SREBP1-driven noncanonical type I IFNs. EMBO Rep. 2024;26:560–592.
- Dang EV. Oxysterol restraint of cholesterol synthesis prevents AIM2 inflammasome activation. Cell. 2017;171(5):1057–1071.
- Widenmaier SB, et al. NRF1 is an ER membrane sensor that is central to cholesterol homeostasis. Cell. 2017;171(5):1094–1109.
- York AG, et al. Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell. 2015;163(7):1716–1729.
- Jethwa C, et al. Control of cholesterol-induced adipocyte inflammation by the Nfe2l1-Atf3 pathway. [preprint]. https://doi.org/10.1101/2024.07.22.604614 Posted on bioRxiv July 23, 2024.
- Zhivaki D, Kagan JC. Innate immune detection of lipid oxidation as a threat assessment strategy. Nat Rev Immunol. 2022;22(5):322–330.
- Yang S, et al. Sugar alcohols of polyol pathway serve as alarmins to mediate local-systemic innate immune communication in drosophila. Cell Host Microbe. 2019;26(2):240–251.
- Okabe Y, Medzhitov R. Tissue biology perspective on macrophages. Nat Immunol. 2016;17(1):9–17.
- Hotamisligil GS, Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nat Rev Immunol. 2008;8(12):923–934.
- Lumeng CN. Innate immune activation in obesity. Mol Aspects Med. 2013;34(1):12–29.
- Denicolai M, et al. Interleukin-1 blockade in patients with ST-segment elevation myocardial infarction across the spectrum of coronary artery disease complexity. J Cardiovasc Pharmacol. 2025;85(3):200–210.
- Kazmi S, et al. The efficacy and safety of interleukin-1 receptor antagonist in stroke patients: A systematic review. J Clin Neurosci. 2024;120:120–128.
- Colaço HG, et al. Tetracycline antibiotics induce host-dependent disease tolerance to infection. Immunity. 2021;54(1):53–67.
- Figueiredo N, et al. Anthracyclines induce DNA damage response-mediated protection against severe sepsis. Immunity. 2013;39(5):874–884.
- Chora AF, et al. DNA damage independent inhibition of NF-κB transcription by anthracyclines. Elife. 2022;11:e77443.
- Passelli K, et al. Strategies for overcoming tumour resistance to immunotherapy: harnessing the power of radiation therapy. Br J Radiol. 2024;97(1160):1378–1390.
- Birnboim-Perach R, Benhar I. Using Combination therapy to overcome diverse challenges of immune checkpoint inhibitors treatment. Int J Biol Sci. 2024;20(10):3911–3922.
- Pradeu T, et al. The conceptual foundations of innate immunity: Taking stock 30 years later. Immunity. 2024;57(4):613–631.
- Newton K, et al. Dying cells fan the flames of inflammation. Science. 2021;374(6571):1076–1080.
- Kroemer G, et al. Immunogenic cell stress and death. Nat Immunol. 2022;23(4):487–500.
- Orzalli MH, et al. Virus-mediated inactivation of anti-apoptotic Bcl-2 family members promotes Gasdermin-E-dependent pyroptosis in barrier epithelial cells. Immunity. 2021;54(7):1447–1462.
- Roncaioli JL, et al. A hierarchy of cell death pathways confers layered resistance to shigellosis in mice. Elife. 2023;12:e83639.
- White MJ, et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159(7):1549–1562.
- Zhang Y, et al. Structural damage in the C. elegans epidermis causes release of STA-2 and induction of an innate immune response. Immunity. 2015;42(2):309–320.
- Zhang H, et al. The extracellular matrix integrates mitochondrial homeostasis. Cell. 2024;187(16):4289–4304.
- Rebendenne A, et al. SARS-CoV-2 predation of Golgi-bound PI4P primes the massive activation of the DNA Damage Response kinase ATM in the cytoplasm [preprint]. https://doi.org/10.1101/2024.12.05.626967 Posted on bioRxiv December 06, 2024.
- Chen J, Chen ZJ. PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation. Nature. 2018;564(7734):71–76.
- Lepelley A, et al. Mutations in COPA lead to abnormal trafficking of STING to the Golgi and interferon signaling. J Exp Med. 2020;217(11):e20200600.
- Uhlorn BL, et al. Attenuation of cGAS/STING activity during mitosis. Life Sci Alliance. 2020;3(9):e201900636.
- Green RS, et al. Mammalian N-glycan branching protects against innate immune self-recognition and inflammation in autoimmune disease pathogenesis. Immunity. 2007;27(2):308–320.
- Martinon F, et al. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11(5):411–418.
- Sweet LA, et al. IRE1α-driven inflammation promotes clearance of citrobacter rodentium infection. Infect Immun. 2022;90(1):e0048121.
- Keestra-Gounder AM, et al. NOD1 and NOD2 signalling links ER stress with inflammation. Nature. 2016;532(7599):394–397.
- Di Conza G, et al. Control of immune cell function by the unfolded protein response. Nat Rev Immunol. 2023;23(9):546–562.
- Aldridge JE, et al. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One. 2007;2(9):e874.
- Romero-Ramirez L, et al. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004;64(17):5943–5947.
- Rouschop KM, et al. PERK/eIF2α signaling protects therapy resistant hypoxic cells through induction of glutathione synthesis and protection against ROS. Proc Natl Acad Sci U S A. 2013;110(12):4622–4627.
- Köditz J, et al. Oxygen-dependent ATF-4 stability is mediated by the PHD3 oxygen sensor. Blood. 2007;110(10):3610–3617.
- Bettigole SE, Glimcher LH. Endoplasmic reticulum stress in immunity. Annu Rev Immunol. 2015;33:107–138.
- Pillich H, et al. Activation of the unfolded protein response by Listeria monocytogenes. Cell Microbiol. 2012;14(6):949–964.
- Grootjans J, et al. The unfolded protein response in immunity and inflammation. Nat Rev Immunol. 2016;16(8):469–484.
- Garfinkel BP, Hotamisligil GS. ER stress promotes inflammation through Re-wIREd macrophages in obesity. Mol Cell. 2017;66(6):731–733.
- Cao S, et al. The IRE1α/XBP1 pathway sustains cytokine responses of group 3 innate lymphoid cells in inflammatory bowel disease. J Clin Invest. 2024;134(13):e174198.
- Fionda C, Sciumè G. A little ER stress isn’t bad: the IRE1α/XBP1 pathway shapes ILC3 functions during intestinal inflammation. J Clin Invest. 2024;134(13):e182204.
- Sullivan GP, et al. TRAIL receptors serve as stress-associated molecular patterns to promote ER-stress-induced inflammation. Dev Cell. 2020;52(6):714–730.
- Wang A, et al. Innate immune sensing of lysosomal dysfunction drives multiple lysosomal storage disorders. Nat Cell Biol. 2024;26(2):219–234.
- Pu X, Qi B. Lysosomal dysfunction by inactivation of V-ATPase drives innate immune response in C. elegans. Cell Rep. 2024;43(5):114138.
- Houtkooper RH, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497(7450):451–457.
- Baker BM, et al. Protective coupling of mitochondrial function and protein synthesis via the eIF2α kinase GCN-2. PLoS Genet. 2012;8(6):e1002760.
- Pellegrino MW, et al. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature. 2014;516(7531):414–417.
- Vesala L, et al. Mitochondrial perturbation in immune cells enhances cell-mediated innate immunity in Drosophila. BMC Biol. 2024;22(1):60.
- West AP, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520(7548):553–557.
- Bronner DN, et al. Endoplasmic reticulum stress activates the inflammasome via NLRP3- and caspase-2-driven mitochondrial damage. Immunity. 2015;43(3):451–462.
- Vazquez C, Horner SM. MAVS coordination of antiviral innate immunity. J Virol. 2015;89(14):6974–6977.
- Lei Y, et al. Elevated type I interferon responses potentiate metabolic dysfunction, inflammation, and accelerated aging in mtDNA mutator mice. Sci Adv. 2021;7(22):eabe7548.
- Sato H, et al. Downregulation of mitochondrial biogenesis by virus infection triggers antiviral responses by cyclic GMP-AMP synthase. PLoS Pathog. 2021;17(10):e1009841.
- Tigano M, et al. Nuclear sensing of breaks in mitochondrial DNA enhances immune surveillance. Nature. 2021;591(7850):477–481.
- Coux O, et al. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847.
- Grover M, et al. Proteasome inhibition triggers tissue-specific immune responses against different pathogens in C. elegans. PLoS Biol. 2024;22(3):e3002543.
- Youngner JS, et al. Influence of inhibitors of protein synthesis on interferon formation in mice. Virology. 1965;27(4):541–550.
- Govindan JA, et al. Lipid signalling couples translational surveillance to systemic detoxification in Caenorhabditis elegans. Nat Cell Biol. 2015;17(10):1294–1303.
- Dunbar TL, et al. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe. 2012;11(4):375–386.
- McEwan DL, Kirienko NV. Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans. Cell Host Microbe. 2012;11(4):364–374.
- Reddy KC, et al. The C. elegans CCAAT-enhancer-binding protein gamma is required for surveillance immunity. Cell Rep. 2016;14(7):1581–1589.
- Fontana MF, et al. Activation of host mitogen-activated protein kinases by secreted Legionella pneumophila effectors that inhibit host protein translation. Infect Immun. 2012;80(10):3570–3575.
- Fontana MF, et al. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 2011;7(2):e1001289.
- Wan L, et al. Translation stress and collided ribosomes are co-activators of cGAS. Mol Cell. 2021;81(13):2808–2822.
- Stein KC, et al. Ageing exacerbates ribosome pausing to disrupt cotranslational proteostasis. Nature. 2022;601(7894):637–642.
- Xu H, et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513(7517):237–241.
- Kufer TA, et al. The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell Microbiol. 2007;10(2):477–486.View this article via: PubMed Google Scholar
- Magalhaes JG, et al. Murine Nod1 but not its human orthologue mediates innate immune detection of tracheal cytotoxin. EMBO Rep. 2005;6(12):1201–1207.
- Acharya D, et al. Actin cytoskeleton remodeling primes RIG-I-like receptor activation. Cell. 2022;185(19):3588–3602.
- Lood C, et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med. 2016;22(2):146–153.
- Zhang W, et al. Targeting KDM4A epigenetically activates tumor-cell-intrinsic immunity by inducing DNA replication stress. Mol Cell. 2021;81(10):2148–2165.
- Zhang G, et al. CPT1A induction following epigenetic perturbation promotes MAVS palmitoylation and activation to potentiate antitumor immunity. Mol Cell. 2023;83(23):4370–4385.
- Riedel CG, et al. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat Cell Biol. 2013;15(5):491–501.
- Li T, et al. Phosphorylation and chromatin tethering prevent cGAS activation during mitosis. Science. 2021;371(6535):eabc5386.
- Kim C, et al. PARP1 inhibitors trigger innate immunity via PARP1 trapping-induced DNA damage response. Elife. 2020;9:1.
- Lu C, et al. DNA sensing in mismatch repair-deficient tumor cells is essential for anti-tumor immunity. Cancer Cell. 2021;39(1):96–108.
- Cheng YC, et al. Topoisomerase I inhibition and peripheral nerve injury induce DNA breaks and ATF3-associated axon regeneration in sensory neurons. Cell Rep. 2021;36(10):109666.
- Mukherjee S, et al. Mechanistic link between DNA damage sensing, repairing and signaling factors and immune signaling. Adv Protein Chem Struct Biol. 2019;115:297–324.
- Komarova EA, et al. p53 is a suppressor of inflammatory response in mice. FASEB J. 2005;19(8):1030–1032.
- Dunphy G, et al. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol Cell. 2018;71(5):745–760.
- Härtlova A, et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42(2):332–343.
- Nowak-Wegrzyn A, et al. Immunodeficiency and infections in ataxia-telangiectasia. J Pediatr. 2004;144(4):505–511.
- Masucci G, et al. Epstein-Barr virus (EBV)-specific cell-mediated and humoral immune responses in ataxia-telangectasia patients. J Clin Immunol. 1984;4(5):369–382.
- Newman LE, et al. Mitochondrial DNA replication stress triggers a pro-inflammatory endosomal pathway of nucleoid disposal. Nat Cell Biol. 2024;26(2):194–206.
- Shen JZ, et al. FBXO44 promotes DNA replication-coupled repetitive element silencing in cancer cells. Cell. 2021;184(2):352–369.
- Chatzidoukaki O, et al. R-loops trigger the release of cytoplasmic ssDNAs leading to chronic inflammation upon DNA damage. Sci Adv. 2021;7(47):eabj5769.
- Maxwell MB, et al. ARID1A suppresses R-loop-mediated STING-type I interferon pathway activation of anti-tumor immunity. Cell. 2024;187(13):3390–3408.
- Mender I, et al. Telomere stress potentiates STING-dependent anti-tumor immunity. Cancer Cell. 2020;38(3):400–411.
- Nassour J, et al. Telomere-to-mitochondria signalling by ZBP1 mediates replicative crisis. Nature. 2023;614(7949):767–773.
- Pokatayev V, et al. RNase H2 catalytic core Aicardi-Goutières syndrome-related mutant invokes cGAS-STING innate immune-sensing pathway in mice. J Exp Med. 2016;213(3):329–336.
- Ishak CA, et al. Spliceosome-targeted therapies induce dsRNA responses. Immunity. 2021;54(1):11–13.
- Bowling EA, et al. Spliceosome-targeted therapies trigger an antiviral immune response in triple-negative breast cancer. Cell. 2021;184(2):384–403.
- Lee-Kirsch MA. Sensing of RNA stress by mTORC1 drives autoinflammation. J Clin Invest. 2022;132(2):e156119.
-
Version history
- Version 1 (August 1, 2025): Electronic publication
- Version 2 (August 4, 2025): Corrected author name in ref. 6
Article tools
-
View PDF

- Download citation information
- Send a comment
- Terms of use
- Standard abbreviations
- Need help? Email the journal
Metrics
Go to
- Top
- Abstract
- Introduction
- Models of immune response initiation
- Surveillance immunity model hypothesis
- Disruption of systemic and metabolic homeostasis
- Disruption of cellular, organellar and molecular homeostasis
- Conclusions and future perspectives
- Acknowledgments
- Footnotes
- References
- Version history



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)





