Citation Information: J Clin Invest. 2012;122(2):654-673. https://doi.org/10.1172/JCI60556.
Abstract
Herpes simplex virus type 1 (HSV-1) not only causes painful recurrent oral-labial infections, it can also cause permanent brain damage and blindness. There is currently no HSV-1 vaccine. An effective vaccine must stimulate coordinated T cell responses, but the large size of the genome and the low frequency of HSV-1–specific T cells have hampered the search for the most effective T cell antigens for inclusion in a candidate vaccine. We have now developed what we believe to be novel methods to efficiently generate a genome-wide map of the responsiveness of HSV-1–specific T cells, and demonstrate the applicability of these methods to a second complex microbe, vaccinia virus. We used cross-presentation and CD137 activation–based FACS to enrich for polyclonal CD8+ T effector T cells. The HSV-1 proteome was prepared in a flexible format for analyzing both CD8+ and CD4+ T cells from study participants. Scans with participant-specific panels of artificial APCs identified an oligospecific response in each individual. Parallel CD137-based CD4+ T cell research showed discrete oligospecific recognition of HSV-1 antigens. Unexpectedly, the two HSV-1 proteins not previously considered as vaccine candidates elicited both CD8+ and CD4+ T cell responses in most HSV-1–infected individuals. In this era of microbial genomics, our methods — also demonstrated in principle for vaccinia virus for both CD8+ and CD4+ T cells — should be broadly applicable to the selection of T cell antigens for inclusion in candidate vaccines for many pathogens.
Authors
Lichen Jing, Jürgen Haas, Tiana M. Chong, Joseph J. Bruckner, Greg C. Dann, Lichun Dong, Joshua O. Marshak, Christopher L. McClurkan, Tori N. Yamamoto, Susanne M. Bailer, Kerry J. Laing, Anna Wald, Georges M.G.M. Verjans, David M. Koelle
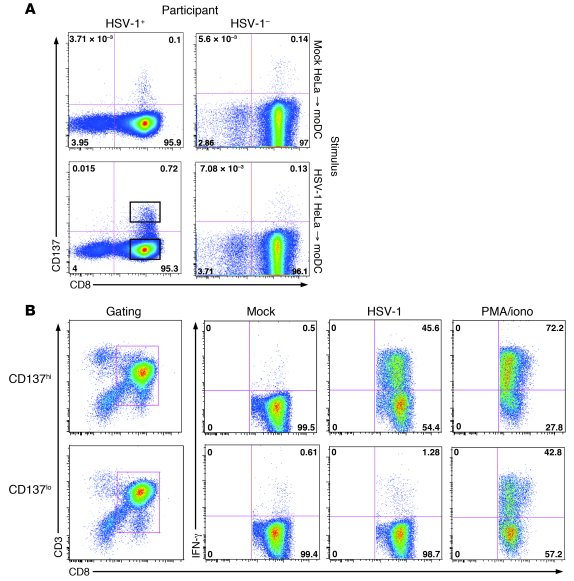
Figure 1
Use of CD137 to detect and enrich HSV-1–specific CD8+ T cells from blood.



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)