Advertisement
Review Series
Open Access |  10.1172/JCI191939
10.1172/JCI191939
KRAS: the Achilles’ heel of pancreas cancer biology
Kristina Drizyte-Miller,1 Taiwo Talabi,2 Ashwin Somasundaram,1,2 Adrienne D. Cox,1,3,4 and Channing J. Der1,3
1Lineberger Comprehensive Cancer Center,
2Department of Medicine,
3Department of Pharmacology, and
4Department of Radiation Oncology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Address correspondence to: Channing J. Der, University of North Carolina at Chapel Hill, Lineberger Comprehensive Cancer Center, Chapel Hill, North Carolina 27599, USA. Email: cjder@med.unc.edu.
Find articles by Drizyte-Miller, K. in: PubMed | Google Scholar
1Lineberger Comprehensive Cancer Center,
2Department of Medicine,
3Department of Pharmacology, and
4Department of Radiation Oncology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Address correspondence to: Channing J. Der, University of North Carolina at Chapel Hill, Lineberger Comprehensive Cancer Center, Chapel Hill, North Carolina 27599, USA. Email: cjder@med.unc.edu.
Find articles by Talabi, T. in: PubMed | Google Scholar
1Lineberger Comprehensive Cancer Center,
2Department of Medicine,
3Department of Pharmacology, and
4Department of Radiation Oncology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Address correspondence to: Channing J. Der, University of North Carolina at Chapel Hill, Lineberger Comprehensive Cancer Center, Chapel Hill, North Carolina 27599, USA. Email: cjder@med.unc.edu.
Find articles by Somasundaram, A. in: PubMed | Google Scholar
1Lineberger Comprehensive Cancer Center,
2Department of Medicine,
3Department of Pharmacology, and
4Department of Radiation Oncology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Address correspondence to: Channing J. Der, University of North Carolina at Chapel Hill, Lineberger Comprehensive Cancer Center, Chapel Hill, North Carolina 27599, USA. Email: cjder@med.unc.edu.
Find articles by Cox, A. in: PubMed | Google Scholar
1Lineberger Comprehensive Cancer Center,
2Department of Medicine,
3Department of Pharmacology, and
4Department of Radiation Oncology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Address correspondence to: Channing J. Der, University of North Carolina at Chapel Hill, Lineberger Comprehensive Cancer Center, Chapel Hill, North Carolina 27599, USA. Email: cjder@med.unc.edu.
Find articles by Der, C. in: PubMed | Google Scholar
Published August 15, 2025 - More info
J Clin Invest. 2025;135(16):e191939. https://doi.org/10.1172/JCI191939.
© 2025 Drizyte-Miller et al. This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
-
Abstract
The genetic landscape of pancreatic ductal adenocarcinoma (PDAC) is well-established and dominated by four key genetic driver mutations. Mutational activation of the KRAS oncogene is the initiating genetic event, followed by genetic loss of function of the CDKN2A, TP53, and SMAD4 tumor suppressor genes. Disappointingly, this information has not been leveraged to develop clinically effective targeted therapies for PDAC treatment, where current standards of care remain cocktails of conventional cytotoxic drugs. Nearly all (~95%) PDAC harbors KRAS mutations, and experimental studies have validated the essential role of KRAS mutation in PDAC tumorigenic and metastatic growth. Identified in 1982 as the first gene shown to be aberrantly activated in human cancer, KRAS has been the focus of intensive drug discovery efforts. Widely considered “undruggable,” KRAS has been the elephant in the room for PDAC treatment. This perception was shattered recently with the approval of two KRAS inhibitors for the treatment of KRASG12C-mutant lung and colorectal cancer, fueling hope that KRAS inhibitors will lead to a breakthrough in PDAC therapy. In this Review, we summarize the key role of aberrant KRAS signaling in the biology of pancreatic cancer; provide an overview of past, current, and emerging anti-KRAS treatment strategies; and discuss current challenges that limit the clinical efficacy of directly targeting KRAS for pancreatic cancer treatment.
-
Introduction
Following lung and colorectal cancer (CRC), pancreatic cancer is the third leading cause of cancer deaths in the United States (1). Whereas recent decades have seen declines in the mortality rates for lung cancer and CRC, pancreatic cancer mortality rates have gradually increased, in part because of the obesity epidemic. Indeed, pancreatic cancer is projected to surpass CRC and become the second leading cause of cancer-related mortality by 2040 (2). Although its 5-year overall survival (OS) rate has improved from 4% in the mid-1990s to 13%, it remains among the lowest of all cancer types (1).
Pancreatic ductal adenocarcinoma (PDAC), an exocrine neoplasm, is the most common subtype of pancreatic cancer, accounting for over 90% of pancreatic neoplasms (3). Despite a well-defined genetic landscape of PDAC (4), no effective targeted therapies have been approved for the majority of patients with PDAC, and the standard of care remains surgery and chemotherapy (5). Most patients are diagnosed with advanced metastatic disease (1), with only 15%–20% eligible for surgery at diagnosis (6). For unresectable PDAC, the first-line therapy is either a combination of 5-fluorouracil, leucovorin, irinotecan, and oxaliplatin, termed FOLFIRINOX (7), or the combination of gemcitabine and nanoparticle albumin-bound paclitaxel (nab-paclitaxel) (8). Disappointingly, the current standard of care is associated with high toxicity, and the median OS on a first-line therapy is less than 12 months and even lower (less than 7 months) on a second-line therapy (7, 8).
The Kirsten rat sarcoma (KRAS) oncogene was identified originally as a retroviral gene responsible for the oncogenic properties of the Kirsten murine sarcoma virus and was later determined to have been transduced from the normal rat genome (Table 1). The discovery of activated KRAS oncogenes in human cancer cell lines in 1982 (9, 10), and their establishment as a sufficient (11) and necessary driver of PDAC growth (12–14), supported the potential significance of KRAS-targeted therapies for PDAC treatment. However, KRAS was initially considered an undruggable cancer target (15). Early efforts focused on indirect strategies to inhibit KRAS membrane association and downstream effector signaling but with minimal therapeutic success (16). It took nearly 40 years until the first direct KRAS inhibitors, targeting a specific mutation (KRASG12C), were clinically approved for non–small cell lung cancer (NSCLC) treatment (17, 18). The successful development of direct KRASG12C inhibitors had a tsunami effect on drug discovery, with more than 50 mutation-selective and pan/multi KRAS/RAS inhibitors now under clinical evaluation (16, 19) (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI191939DS1).
In this Review, we focus on KRAS as the Achilles’ heel of pancreatic cancer treatment. It is both the critical driver of PDAC growth as well as arguably the greatest therapeutic vulnerability for PDAC treatment. We revisit the early indirect strategies of targeting KRAS and provide an overview of the current landscape of direct KRAS inhibitors. We end with a discussion of lessons learned from the results from ongoing clinical trials, resistance mechanisms to KRAS inhibitors, and potential combination strategies to improve outcomes for patients with pancreatic cancer.
-
KRAS — the driver of pancreatic cancer
Molecular and histological profiling demonstrated that approximately 85%–90% of PDAC is initiated from the precursor lesions, termed pancreatic intraepithelial neoplasms (PanINs) (Figure 1A), with the remaining 10%–15% arising from mucinous pancreatic cyst precursors, most often intraductal papillary mucinous neoplasms (IPMNs) (20, 21). These precursor lesions undergo a stepwise accumulation of gain- and loss-of-function genetic mutations as they progress to invasive and metastatic PDAC (22).
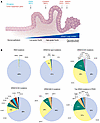 Figure 1
Figure 1KRAS mutations in PDAC. (A) Schematic illustrating pancreatic ductal adenocarcinoma (PDAC) pathogenesis and progression (adapted from ref. 176 with permission from Springer Nature Limited, which retains the rights to the reference image). Mutations in KRAS oncogene are the initiating step in PDAC development, and they induce transformation of normal pancreas epithelium to low-grade pancreatic intraepithelial neoplasia (PanIN). Progression from low-grade PanINs to high-grade PanINs and eventually invasive PDAC is caused by loss-of-function mutations in CDKN2A, TP53, and SMAD4 tumor suppressor genes. The severity of disease is also associated with increased KRASmut copy numbers. (B) KRAS mutation frequencies in PDAC. Data were compiled from the cBioPortal GENIE Cohort v17.0 database (48) from 7,407 patients with PDAC. Of the three RAS isoforms, KRAS is the predominantly mutated isoform, with NRAS and HRAS mutations accounting for <1% of PDAC cases. Of the three mutational hot spots, G12X mutations are most common in PDAC, with G12D, G12V, and G12R representing the predominant amino acid mutations at this position. G13X mutations are rare in PDAC and comprise less than 1% of KRAS mutations. Q61X mutations are also uncommon, accounting for 7% of KRAS point mutations, with Q61H representing the predominant mutation. The authors would like to acknowledge the American Association for Cancer Research and its financial and material support in the development of the AACR Project GENIE registry, as well as members of the consortium for their commitment to data sharing. Interpretations are the responsibility of the authors.
Genome-wide sequence analyses have identified four predominant gene mutations in PDAC (23–31). In addition to gain-of-function KRAS missense mutations, loss-of-function mutations in CDKN2A (with additional loss mediated by homozygous deletion or promoter hypermethylation), TP53, and/or SMAD4 tumor suppressor genes dominate the genetic landscape of PDAC (Figure 1A). Genetic profiling of early-stage preneoplastic lesions supports a model in which mutations in these genes contribute to the initiation of neoplasia and progression to invasive and metastatic PDAC (4). While the cell of origin of PDAC is still debated, genetic studies in mouse models support development of an acinar-to-ductal metaplasia in the epithelia of the exocrine pancreas upon acquisition of oncogenic mutations in Kras (32). Recent clinicogenomic profiling of 2,336 tumors from patients with both resectable and metastatic PDAC found KRAS mutations in 95% of cases, followed by TP53 mutation in 76%, CDKN2A/B mutation in 38%, and SMAD4 mutation in 24% of cases (33).
Genetically engineered mouse models (GEMMs) have been instrumental tools in establishing KRAS mutations as the initiating event in PDAC tumorigenesis (34, 35) (Table 1). Conditional expression of KrasG12D in pancreatic progenitor cells was sufficient to induce the formation of PanIN lesions that histologically recapitulated the PanIN stages observed in human PDAC, characterized by long latencies and low penetrance of invasive and metastatic disease (11). However, when mutant Kras was combined with inactivation of the Trp53 (36), Ink4a/Arf (37), or Smad4 (38) tumor suppressor genes, it resulted in rapid progression of PanINs and fully penetrant development of invasive and highly metastatic PDAC.
The role of KRAS mutations as the critical initiating genetic step in PDAC is also supported by genetic profiling of PanIN lesions, which are characterized as low-grade (LG) and high-grade (HG) PanINs (39) (Figure 1A). KRAS but not tumor suppressor mutations are found in LG PanINs (40, 41). Deletions and mutations in CDKN2A (encoding p16INK4a and p14ARF) are found in LG PanINs and increase in frequency in HG PanINs, with additional loss of CDKN2A expression mediated by promoter hypermethylation (42). TP53 mutations are found in HG PanINs and increase in frequency in PDAC, whereas SMAD4 mutations are found in advanced PDAC. KRAS mutations are already present in 95% of LG and HG PanIN lesions, consistent with their initiating role (40, 41, 43). However, LG and HG PanIN lesions are present in cancer-free elderly individuals, indicating that additional genetic steps are essential to unleash the oncogenic driver function of mutant KRAS (44).
GEMMs have been key in establishing that mutant KRAS is also necessary for tumor maintenance (Table 1). Specifically, inactivation of mutant KrasG12D in early-stage or established PanINs caused regression of primary (12, 14, 16) and metastatic (13) tumors. Similarly, genetic silencing by RNA interference (RNAi) in KRAS-mutant human cancer cell lines also caused growth suppression (45, 46). Together, these findings support targeting KRAS as a therapeutic approach in advanced PDAC.
-
KRAS mutations
Approximately 20% of all human cancers harbor RAS mutations (47); KRAS is the most frequently mutated isoform (83%), followed by NRAS (15%), with HRAS mutated infrequently (2%) (GENIE Cohort v17.0; ref. 48). RAS mutations are seen predominantly at one of three mutational hot spots: glycine-12 (G12), glycine-13 (G13), and glutamine-61 (Q61). The frequency of specific RAS gene mutations is highly skewed, with the majority of cancers predominantly expressing a mutation of one specific RAS allele (49–51). PDAC is characterized by mutations near-exclusively in KRAS (99%), with HRAS and NRAS mutations occurring in 0.7% and 0.3% of patients with PDAC, respectively (GENIE Cohort v17.0; ref. 48) (Figure 1B). Most missense mutations in KRAS occur at the G12 (91%) and Q61 (7%) amino acid positions, with G13 mutations being rare (1%). G12D is the most frequent substitution (41%), followed by G12V (32%) and G12R (16%). This mutation profile is nearly identical to that seen in PanIN lesions (43, 52), further supporting KRAS mutation as the initiating genetic event in PDAC.
The prominence of G12R mutations in PDAC contrasts strikingly with other cancers that harbor high levels of KRAS mutations; G12R mutations are found in only 1%–2% of NSCLC and CRC (16). Conversely, the smoking-associated KRASG12C mutation is the most prevalent KRAS mutation in NSCLC (40%), but it is found in less than 2% of PDAC (53). G13X mutations comprise 18% of KRAS mutations in CRC, yet they represent less than 1% of KRAS mutations in PDAC. The basis for mutation of a specific RAS gene or for mutation at specific hot spots in different cancer types remains poorly understood (54). There is evidence for both DNA mutagenic mechanisms as well as biological properties as influencing these frequencies (55, 56). This topic has been addressed in considerable detail in other recent reviews (50, 54, 57).
An emerging concept is that different mutations have distinct consequences for KRAS oncogenic function and, consequently, may exhibit differential therapeutic vulnerabilities (57). Evaluation of patients with PDAC indicates that the different KRAS mutations are associated with different clinical characteristics. KRASG12D has been associated with the worst survival, whereas KRASG12R has been associated with improved survival (33, 52). Compared with KRASG12D, KRASG12R mutant PDAC also has a less invasive phenotype, enriched in early-stage (stage I) versus late-stage (stage II–III) disease (44% versus 24%), and diminished metastatic potential, with increased lymph node negativity (47% versus 26%). These clinical differences suggest that KRASG12R is a less potent cancer driver and are consistent with preclinical GEMM analyses where KrasG12R did not effectively drive PanIN formation (58). A possible mechanistic basis for the reduced oncogenic potency of KRASG12R may be based in part on reduced migration potential (52) as well as impaired phosphoinoside-3-kinase (PI3K) effector activation and promotion of macropinocytosis (59), a metabolic activity essential for PDAC tumorigenicity (60).
Gene dosage has also emerged as an important parameter that may support mutant KRAS driver function and impact the clinical disease and responses to therapy (33). Increased copy numbers of mutant versus WT KRAS alleles were associated with worse OS in both resectable (23 months versus 32 months) and metastatic (8.5 months versus 13 months) disease. Mechanisms that increase KRAS mutant copy number include preferential amplification of the mutant allele and loss of the WT allele. Whole-genome duplication, which is seen in nearly two-thirds of patients (25), also enhances mutant allele copy numbers. KRAS gene amplification is also associated with acquired resistance to direct KRAS inhibitors (16). Finally, GEMM studies support a potential tumor suppressor function of the KRAS WT allele (61–63). Consistent with this, loss of the WT allele in patients with KRAS mutant copy gains has been associated with significantly worse OS (33). These observations should be taken into consideration when prioritizing the development of therapeutic approaches for patients harboring a particular KRAS allele mutation or amplification. The development of highly potent inhibitors against the three most prevalent KRAS mutations in PDAC (G12D, G12V, and G12R), along with agents that would sustain pathway inhibition in KRAS-amplified tumors, might yield the most impactful therapeutic benefit for patients with PDAC.
Among the 5% of PDAC that are KRAS WT, 60% exhibit genetic alterations in the upstream receptor tyrosine kinases (RTKs) (e.g., NTRK1, NTRK3, FGFR2, ERBB2, ROS1, and MET), at the level of RAS or RAS regulation (e.g., NRAS, NF1) or in components of the downstream RAF/MEK/ERK MAPK cascade (e.g., BRAF, RAF1 and MAP2K1 (which encodes MEK1) (33). The remaining 40% of KRAS WT PDAC lack mutations in the ERK MAPK signaling network and instead are enriched in GNAS, SMARCB1, and PIK3CA mutations. KRAS WT PDAC, with and without other ERK MAPK network mutations, exhibits improved OS and response to therapy (33, 52).
-
KRAS signaling in PDAC
KRAS encodes two highly similar (~85% amino acid identity) isoforms (KRAS4A and KRAS4B) that are produced by alternative splicing of exon four and differ solely in their carboxyl-terminal residues (64) (Figure 2A). KRAS is a small GTPase that functions as a binary on-off switch that relays extracellular signal-induced stimuli to cytoplasmic signaling networks. It comprises an amino-terminal catalytic G domain responsible for binding and hydrolyzing GTP to GDP and a carboxyl-terminal hypervariable region (HVR), which undergoes posttranslational lipid modifications critical for membrane targeting (65–68).
 Figure 2
Figure 2KRAS GTPase regulation and signaling. (A) KRAS encodes a small GTPase comprising the G domain and hypervariable region (HVR). Alternative splicing of exon four results in two KRAS isoforms (KRAS4A/KRAS4B, denoted as 4A/B), which differ in their carboxyl-terminal 151–188/189 amino acids. The G domain is involved in guanosine triphosphate (GTP) and guanosine diphosphate (GDP) binding and interactions with guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs), and effectors. HVR contains the CAAX tetrapeptide motif that acts as a signal for posttranslational modifications that promote KRAS plasma membrane association essential for KRAS oncogenic function. Switch I and II regions (denoted as SI and SII) are highlighted, and mutational hot spots at G12, G13, and Q61 positions are indicated with red asterisks. (B) KRAS cycles between active GTP-bound and inactive GDP-bound states. Receptor tyrosine kinase (RTK) signaling promotes GEF-mediated GTP loading and activation of KRAS, which then engages downstream effector signaling (i.e., the RAF/MEK/ERK MAPK cascade). GAPs accelerate intrinsic KRAS GTPase activity and GTP hydrolysis to return KRAS to the inactive GDP-bound state. Amino acid substitutions at G12, G13, and Q61 hot spot positions accelerate GDP to GTP exchange rates and/or impair intrinsic or GAP-induced GTP hydrolysis, resulting in constitutively active KRAS. (C) KRAS undergoes three posttranslational modifications at the carboxyl-terminal CAAX motif (where C denotes cysteine, A denotes aliphatic, and X denotes terminal residues), which is required for association with membranes. Farnesyltransferase (FTase) adds a 15-carbon farnesyl group to the cysteine amino acid at the CAAX motif, RAS-converting enzyme (RCE1) removes -AAX residues, and isoprenylcysteine carboxylmethyltransferase (ICMT) catalyzes carboxylmethylation of farnesylated cysteine. Inhibition of FTase (FTIs) leads to alternative prenylation of KRAS by geranylgeranyltransferase-I (GGTase-I), which adds a 20-carbon geranylgeranyl group and facilitates KRAS associate with membranes. C, cysteine; Ome, carboxyl methylation.
The intrinsically low GTP hydrolysis and exchange activities of WT KRAS are accelerated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs), respectively (69, 70) (Figure 2B). GEFs (e.g., SOS1) assist in GTP/GDP exchange while GAPs (e.g., NF1) facilitate hydrolysis of the bound GTP. Cycling between the GTP-bound on-state (ON) and GDP-bound off-state (OFF) causes conformational changes in the switch I (residues 30–40) and switch II (residues 60–76) regions of KRAS that are responsible for effector binding and interaction with GEFs and GAPs, respectively (68). The mutational hot spots in KRAS occur near the switch regions and, to varying degrees, reduce both intrinsic and GAP-induced GTP hydrolysis and/or increase intrinsic GDP/GTP exchange rates, both of which favor formation of the constitutively ON KRAS (71).
Canonically, KRAS is activated in response to extracellular stimuli to promote cell growth and survival (Figure 2B). Growth factor–mediated RTK (e.g., EGFR) signaling leads to activation of KRAS and subsequent association with downstream effectors to initiate a multitude of signaling pathways. Although more than 12 functional classes comprising >50 validated/putative RAS effectors have been identified (72, 73), the RAF/MEK/ERK MAPK and the PI3K/AKT/mTOR signaling networks comprise the two best validated effector signaling networks that support KRAS-driven oncogenesis (16).
GTP-bound KRAS, whether WT or mutant, promotes activation of RAF serine/threonine kinases (ARAF, BRAF, and RAF1/CRAF) by a complex mechanism involving relief of autoinhibition, promotion of membrane association, phosphorylation by membrane-associated protein kinases, and dimerization. Activated RAF then phosphorylates and activates the MEK1/2 dual-specificity protein kinases, which then phosphorylate and activate the ERK1/2 serine/threonine kinases (74, 75). In contrast to the limited substrates of RAF and MEK, activated ERK regulates a complex and dynamic phosphoproteome in KRAS-mutant PDAC cells comprising over 2,000 cytoplasmic and nuclear proteins (76). ERK substrates include transcription factors, kinases, epigenetic regulators, E3 ligases, and phosphatases, which in turn alter gene transcription and cellular signaling to promote cell cycle progression and cell growth (77). One of the key downstream substrates of ERK is the transcription factor MYC (76, 77), which regulates tumor metabolism (78) and is critical for KRAS-mutant PDAC growth (79–81).
A second major effector of KRAS is PI3K, which converts phosphatidylinositol-4,5-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3) to activate the AKT1-3 serine/threonine protein kinases (82). Activated AKT then leads to activation of the kinase mTOR and regulation of cell metabolism, proliferation, migration, and survival (83). PI3K signaling has been shown to be essential for KRAS-driven tumorigenesis in vivo, where mutations in Pik3ca (encoding the p110α subunit of PI3K) that result in defective binding to RAS also prevent Kras-driven NSCLC formation and maintenance (84, 85).
Several key findings suggest that the RAF/MEK/ERK MAPK signaling cascade is the major effector of KRAS-driven progression and growth of PDAC. First, activating mutations in Braf, but not Pik3ca, in mice phenocopied Kras mutations in driving PDAC initiation and maintenance when coupled with loss of Tp53 (86). Second, the KRAS-dependent transcriptome and phosphoproteome were nearly identical to the ERK-regulated transcriptome and phosphoproteome in PDAC cells (76, 77). Third, constitutive activation of MEK1 and ERK1/2, but not AKT, rescued KRAS inhibitor-induced growth suppression in PDAC (77). However, mutations in components of the PI3K/AKT/mTOR pathway have been identified in patients who relapsed on KRASG12C inhibitor treatment (87, 88), suggesting that the role of PI3K and other KRAS effectors in KRAS-mutant PDAC needs to be further investigated.
-
Early approaches for anti-KRAS therapies in PDAC
Farnesyltransferase inhibitors. Initial attempts to directly drug KRAS by developing GTP analogs that would compete with GTP binding or GAP-like molecules that would restore the intrinsic GTPase activity were not successful. Instead, the focus shifted toward targeting the farnesyltransferase (FTase) enzyme responsible for adding a 15-carbon farnesyl lipid modification to the carboxyl-terminus of KRAS (Figure 2C). This modification was shown to be required for KRAS association with plasma membrane and for downstream signaling and cell transformation (49). However, FTase inhibitors (FTIs) had disappointing clinical outcomes with no significant efficacy in KRAS-mutant PDAC (89–91) (Table 2). In retrospect, these negative outcomes were predicted by earlier experimental studies that found that FTIs were effective against HRAS- but not KRAS-transformed rodent fibroblasts (92). The explanation for this distinction was that, when FTase activity is blocked, KRAS but not HRAS undergoes alternative prenylation by the FTase-related enzyme, geranylgeranyltransferase-I (GGTase-I), which adds a 20-carbon geranylgeranyl lipid modification to the carboxyl-terminus of KRAS (93, 94) (Figure 2C). This unexpected property of KRAS (and NRAS) was missed by initial studies that focused on HRAS-mutant cell models (92, 95, 96), when it was widely believed that the three RAS proteins were identical in biochemical properties and function. Although therapeutic strategies focused on inhibiting KRAS membrane association are still being pursued, these indirect strategies will likely be limited by additional effects on the functions of non-RAS targets.
Targeting KRAS effector signaling. With over 100 approved oncology drugs, protein kinases are among the most successful class of anticancer targets (97, 98). Thus, the discovery that the ERK/MAPK cascade is a key effector of KRAS-driven cancer growth fueled a second major approach of indirectly targeting KRAS (Table 2). Multiple small-molecule inhibitors of each node of the RAF/MEK/ERK MAPK cascade have been developed and have shown promise in preclinical studies (99, 100). RAF and MEK inhibitors have been approved for BRAF-mutant melanoma and other cancers (101), and one MEK inhibitor has been approved for NF1-deficient plexiform neurofibromas (102). However, the use of ERK MAPK inhibitors for the treatment of PDAC and other KRAS-mutant cancers has been challenging owing to on-target toxicity, acquired resistance, and loss of ERK-dependent negative feedback loops, which ultimately cause reactivation of ERK signaling through RTKs and WT KRAS (74, 75). Although there was some indication of clinical efficacy, clinical evaluation of the ERK-selective inhibitor ulixertinib in PDAC was terminated due to toxicity (103). Similarly, PI3K/AKT/mTOR pathway inhibitors alone or in combination with chemotherapy or other targeted inhibitors showed limited clinical success (82). Despite promising preclinical data using PDAC cell lines and GEMMs, these inhibitors did not demonstrate significant antitumor effects and/or were associated with dose-limiting toxicities in patients with PDAC (104).
Additionally, oncogenic KRAS effector signaling reprograms tumor metabolism in ways that could be exploited for therapeutic benefit (105). KRAS-mutant cancer cells exhibit increased glycolytic flux and increased dependency on glutamine metabolism and on nutrient-scavenging pathways such as autophagy and macropinocytosis, among others (106). These findings have sparked intense interest in targeting metabolic adaptations of KRAS-mutant PDAC, although so far with limited clinical benefit. Several clinical trials have been completed or are ongoing to target autophagy using hydroxychloroquine in combination with chemotherapy (NCT01978184) (107) or with MEK/ERK inhibitors in PDAC (NCT04386057, NCT03825289, NCT04132505) (108, 109). Devimistat (CPI-613), an inhibitor of the tricarboxylic acid cycle, has been evaluated in combination with modified FOLFIRINOX; however, it did not improve outcomes for patients with PDAC compared with chemotherapy alone (NCT01835041) (110). Devimistat is currently being evaluated as a triple combination with hydroxychloroquine and chemotherapy in patients with PDAC (NCT05733000). The glutaminase 1 inhibitor telaglenastat (CB-839) has shown limited clinical efficacy in advanced solid tumors when combined with PARP inhibitors (NCT03875313), perhaps owing to rapid metabolic adaptations that overcome glutamine dependency (111). Greater efficacy will require the development of more tolerable KRAS effector pathway inhibitors (or the use of direct KRAS inhibitors as discussed below) and more effective and selective inhibitors of metabolic pathways (112, 113).
-
Development of direct KRAS inhibitors
KRASG12C inhibitors. KRAS has long been viewed as an “undruggable” target due to its high affinity for GTP (114) and the lack of suitable binding pockets for drug candidates (115). However, Shokat and colleagues challenged this notion in 2013 with the seminal discovery of a previously unseen switch II pocket that became visible only after being stabilized due to its occupancy by a small molecule covalently bound to the cysteine residue at the G12 position (116, 117). Just a few years after the initial discovery of the switch II pocket, two KRASG12C inhibitors, sotorasib (AMG 510) and adagrasib (MRTX849), entered clinical evaluation for KRASG12C-mutant solid tumors (118, 119). The first clinical trial showed that sotorasib had an acceptable safety profile and demonstrated clinical benefit in patients with NSCLC, with an objective (or overall) response rate (ORR) of 37.1% and a median OS of 12.5 months (18, 120). The second clinical trial, of adagrasib, had similar outcomes in NSCLC, with an ORR of 42.9% and a median OS of 12.6 months (17).
Sotorasib and adagrasib were granted accelerated FDA approval for advanced NSCLC in 2021 and 2022, respectively. Furthermore, randomized, open-label phase III trial results demonstrated that the ORR was higher in patients with NSCLC treated with sotorasib compared with the standard-of-care docetaxel (28.1% and 13.2%, respectively), although the median OS was not significantly different between treatments (10.6 months for sotorasib and 11.3 months for docetaxel) (121). Similarly, the phase III trial comparing adagrasib with docetaxel found that the ORR was substantially higher in adagrasib-treated patients with NSCLC (31.9%) compared with docetaxel-treated patients (9.2%). Considering their selectivity for mutant over WT KRAS, these inhibitors caused unexpectedly high levels of treatment-related adverse events (TRAEs) of grade 3 or higher (33% for sotorasib [121], 47% for adagrasib [122]).
Although these initial findings sparked excitement in the KRAS-mutant cancer field, less than 2% of patients with PDAC harbor KRASG12C mutations (Figure 1B), limiting how beneficial this therapeutic avenue might be. KRASG12C-mutant patients with PDAC showed an ORR of 21.1% and median OS of 6.9 months without significant adverse events (123). Slightly better results were observed with adagrasib with ORR of 33.3% and median OS of 8 months (124) (Table 3). Although the response to KRASG12C inhibitors did not outperform the current standard of care, TRAEs were lower after KRASG12C inhibitor treatment compared with chemotherapy (7, 123).
There are now over 20 additional direct KRASG12C inhibitors under clinical evaluation (Supplemental Table 1); the majority target GDP-bound KRASG12C and share a similar mechanism of action to sotorasib and adagrasib (Figure 3). Among these, divarasib has shown potentially superior activity versus the approved inhibitors and is currently in phase III evaluation compared directly with the two approved inhibitors (125). In contrast to the KRASG12C(OFF) inhibitors, BBO-8520 is a first-in-class covalent KRASG12C inhibitor that binds to both GDP- and GTP-bound KRASG12C and is under phase I clinical evaluation in NSCLC (NCT06343402) (126). Additionally, RMC-4998 and its clinical analog elironrasib/RMC-6291 are members of a unique class of KRASG12C inhibitors, where the compound first forms a binary complex with a cytoplasmic chaperon cyclophilin A (CypA) and then binds to GTP-bound KRASG12C, forming a tri-complex (127). Downstream KRAS signaling is inhibited because this tri-complex inhibitor sterically prevents effector interaction with KRAS. Elironrasib is also in phase I clinical trials for advanced KRASG12C solid tumors as a monotherapy (NCT05462717) and in combination with a multi-RAS inhibitor daraxonrasib/RMC-6236 (NCT06128551). There is also evidence to suggest that drug-modified KRASG12C oncoprotein fragments could harness an immune response. Recent proof-of-principle experiments suggested that ARS-1620- (128) or sotorasib-modified KRASG12C (129) are presented as neoantigens by class I MHC, which then recruit cytotoxic T cells to KRASG12C inhibitor-resistant cancer cells. It remains to be determined if any of the newer KRASG12C inhibitors will elicit stronger responses in KRASG12C-mutant PDAC.
 Figure 3
Figure 3Direct KRAS inhibitors. (A) The current landscape of direct KRAS inhibitors and their status in preclinical and clinical stages. Blue indicates approved drugs, green indicates clinical trials that are recruiting, gray indicates active clinical trials that are not recruiting, red indicates terminated clinical trials, and purple indicates trials with unknown status. (B) The mechanisms of action of KRAS inhibitors are diverse. Mutant-selective inhibitors can be off-state, on-state, or off- and on-state inhibitors. Some inhibitors covalently modify mutant KRAS, others do not. There are multi-mutant or pan-KRAS and pan-RAS inhibitors that target WT and mutant KRAS/RAS proteins. Tri-complex inhibitors utilize cytosolic cyclophilin A (CypA) scaffold and KRAS degraders utilize ubiquitin-mediated proteasomal degradation of KRAS protein.
KRASG12D inhibitors. The substantial progress and success of KRASG12C inhibitors has stimulated intense efforts to develop inhibitors against other KRAS mutant proteins. This is of particular relevance to PDAC, where 41% of tumors are driven by KRASG12D mutations (Figure 1B). The first KRASG12D-selective inhibitor, MRTX1133, demonstrated near 1,000-fold selectivity for inhibiting KRASG12D signaling and KRASG12D-mutant cancer cell growth as compared with KRAS WT (130, 131). MRTX1133 exhibited excellent antitumor efficacy and tumor regression, elicited an immune response in preclinical models, and entered phase I/II clinical evaluation for KRASG12D solid tumors in 2023 (130, 132). However, clinical evaluation of MRTX1133 (NCT0537706) was recently terminated because the drug exhibited high pharmacokinetic variability and failed to meet thresholds for advancement.
In contrast to MRTX1133, zoldonrasib/RMC-9805 is a covalent ON KRASG12D-selective inhibitor. Zoldonrasib, a tri-complex inhibitor with CypA, binds to KRASG12D in its GTP-bound state and has demonstrated promising antitumor efficacy as both monotherapy and in combination with anti-PD1 therapy in preclinical KRASG12D models, including PDAC (133). Early clinical evaluation showed promising efficacy in PDAC (30% ORR) with very limited toxicity (Table 3). Several additional OFF (LY3962673, ref. 134, and QTX3046, ref. 135) and ON (GFH375/VS-7375, ref. 136; HRS-4642, ref. 137; TSN1611, ref. 138; and INCB161734, ref. 139) KRASG12D-selective inhibitors are in phase I clinical evaluation (Supplemental Table 1).
Pan-KRAS and multi-KRAS inhibitors. Unlike the allele-selective KRAS inhibitors, pan-KRAS and multi-KRAS inhibitors inhibit multiple KRAS mutants as well as WT KRAS protein (Figure 3). Preclinical compounds BI-2865 and BI-2493 bind to a broad range of GDP-bound mutant KRAS proteins and WT KRAS but not WT HRAS or NRAS (140). In mice bearing KRASG12C/D/V and KRASA146V tumors, BI-2493 has demonstrated antitumor activity and inhibition of ERK phosphorylation without toxicity, as measured by changes in body weight. Although BI-2865 and BI-2493 are considered “pan-KRAS” inhibitors that target 18 of 24 most common KRAS mutations, they lack activity against KRASG12R, KRASQ61L/K/R, and KRASA59T mutant proteins (140). These compounds have also shown activity in KRAS WT-amplified tumors, most commonly seen in gastric and esophageal cancers (141). A clinical candidate BI 3706674 is now under clinical evaluation in cancers harboring KRASG12V or KRAS WT amplifications (NCT06056024). QTX3034, a noncovalent multi-KRAS inhibitor against GDP-bound KRASG12D and to a lesser extent KRASG12V (142), is also under clinical evaluation as a monotherapy or in combination with cetuximab (NCT06227377) for patients with KRASG12D solid tumors (Supplemental Table 1).
Pan-RAS inhibitors. Based on GEMM studies that observed deleterious consequences caused by genetic ablation of Ras genes (143–147), it was anticipated that a pan-RAS inhibitor would be toxic. Therefore, an unexpected and most encouraging clinical development in the field of direct KRAS inhibitors for PDAC treatment is the tri-complex, pan-RAS, ON selective inhibitors. RMC-7977 and its clinical analog daraxonrasib/RMC-6236 are first-in-class reversible tri-complex RAS(ON) pan-RAS-selective inhibitors that bind to both WT and mutant KRAS, NRAS, and HRAS proteins (148–150) (Figure 3). These inhibitors block RAS signaling by preventing effector binding and/or by stimulating intrinsic RAS GTPase activity (151). RMC-7977 treatment demonstrated potent inhibitory activity against a broad spectrum of RAS mutations, with KRASG12X-mutant cancer cell lines displaying the highest degree of sensitivity. Furthermore, RMC-7977 caused robust and durable tumor suppression and multiple regressions in a large panel of KRASG12X PDAC, CRC, and NSCLC xenograft models (148). Recent reports indicated that WT RAS and upstream RTK signaling limit the therapeutic efficacy of KRASG12C inhibitors (152). Due to its ability to bind and inhibit WT RAS proteins, RMC-7977 retained activity in KRASG12C inhibitor-resistant cancer cells (148).
Daraxonrasib is under clinical evaluation in KRAS-mutant solid tumors, including PDAC, with encouraging patient outcomes (NCT05379985). Preliminary reports from 42 patients with PDAC harboring KRASG12X mutations demonstrated a median progression-free survival of 8.5 months and ORR of 27% (Table 3). Importantly, daraxonrasib was well tolerated, and the most common TRAEs were grade 1 or 2 rash, nausea, and vomiting (153). Recruitment for the RASolute 302 phase III clinical trial comparing daraxonrasib as a second-line treatment versus chemotherapy is currently ongoing (NCT06625320).
-
Other KRAS therapeutic strategies
Although the field has been dominated by direct KRAS small-molecule inhibitors, several alternative anti-KRAS strategies, including RNAi, proteolysis targeting chimera (PROTAC), and immunotherapy-based approaches have been under clinical evaluation for KRAS-mutant solid tumors, including PDAC, albeit with less exciting results (Supplemental Table 1). siG12D-LODER is a novel bio-degradable polymeric matrix containing RNAi against KRASG12D/V that is implanted directly into the pancreas. Reports from a phase I/IIa clinical trial demonstrated that combination treatment with siG12D-LODER and chemotherapy was safe and well tolerated with a median OS of 15.1 months, although the current status of this RNAi therapy is unknown (NCT01676259) (154).
The development of PROTAC-based KRAS degraders is another emerging strategy for targeting KRAS-mutant cancers. ASP3082 is a PROTAC degrader that tags mutant KRASG12D for ubiquitin-mediated proteasomal degradation, with a strong selectivity for mutant KRAS protein over >9,000 other proteins. ASP3082 treatment decreased KRASG12D downstream signaling and cancer cell growth in vitro and in xenograft models after once-weekly intravenous administration (155). It is currently in phase I trials and has so far demonstrated an acceptable safety profile in patients with PDAC (NCT05382559) (156). Similarly, ACBI3 is a pan-KRAS PROTAC degrader active against 13 of 17 of the most common KRAS mutations; it demonstrated potent and durable inhibition of KRAS signaling in vitro and tumor regression in vivo (157). There are both advantages and disadvantages of utilizing PROTAC-based degraders compared with small-molecule inhibitors. Degraders might allow for targeting multiple KRAS mutations simultaneously and result in inhibition of all KRAS functions, not solely inhibition of downstream effector binding. However, due to their large molecular size, delivery of PROTACs is challenging and will require intravenous administration, compared with oral delivery of small molecules. It is also unknown if PROTAC degraders will be susceptible to the same or novel resistance mechanisms as small-molecule inhibitors.
Finally, several attempts have been made to use vaccines and T cell therapies to target KRAS. The TG01 vaccine, consisting of synthetic peptides against seven of the most common KRAS mutations, was used in combination with recombinant human GM-CSF. When given with gemcitabine, TG01 evoked an immune response and led to a median OS of 33.3 months, but it is no longer in active development (NCT202261714) (158). Furthermore, mRNA-5671/V94 (159), a lipid nanoparticle-based mRNA vaccine against several KRAS mutations (G12D, G12V, G13D, and G12C), was under clinical evaluation, but the trial has been terminated (NCT03948763). ELI-002 2P is a lymph node–targeted KRASG12D/G12R amphiphile vaccine. Early results demonstrated that ELI-002 2P elicited a notable T cell response without dose-limiting toxicities in patients with PDAC and CRC (NCT04853017) (160). KISIMA-02, another experimental approach under clinical investigation, is a three-component platform consisting of a vaccine against KRASG12D/G12V (ATP150/ATP152), a viral vector (VSV-GP154), and the immune checkpoint inhibitor ezabenlimab (161) (NCT05846516). Recently, trial results (NCT03592888) from a mature dendritic cell vaccine against KRASMUT (mDC3/8-KRAS) were published, which reported a KRASG12V-specific T cell response in vaccinated individuals (162). There is also preliminary evidence that individualized mRNA neoantigen vaccines administered in combination with anti–PD-L1 inhibitors and chemotherapy in surgery-eligible patients with PDAC elicited T cell responses and correlated with delayed recurrence (NCT04161755) (163). Harnessing the immune system as an anti-KRAS therapy has the potential for long-lasting benefits; however, it has so far been largely unsuccessful in the clinic and requires better understanding of the immunosuppressive tumor microenvironment of PDAC.
-
Resistance to KRAS inhibitors and promising combination strategies
The challenge of nearly all targeted therapies is primary (innate) and acquired resistance. Our understanding of resistance mechanisms to KRAS inhibitors in PDAC remains limited and stems primarily from patients with NSCLC, CRC, and PDAC who have been treated with KRASG12C-selective inhibitors (87, 88, 164–167). Unlike the resistance to protein kinase inhibitors, which commonly arises due to second site mutations that impair inhibitor binding, putative KRAS inhibitor resistance mechanisms are varied and complex (Figure 4A). Strikingly, up to a dozen distinct mutations have been found within one patient. Targeted DNA sequencing analyses of circulating tumor DNA in patients experiencing relapse have identified genetic alterations at three distinct levels that ultimately converge to reactivate KRAS signaling. These alterations occurred at the level of RAS itself, in the components upstream of RAS, or in the effectors downstream of RAS (87, 88, 164–169) (Figure 4B). In addition, mutations in components outside the RAS signaling network have also been described (Supplemental Figure 1). Nongenetic mechanisms of resistance were found in half of patients who relapsed on KRASG12C inhibitor treatment (Figure 4C). Some of these include transcriptional reprogramming that changes cellular states such as epithelial-to-mesenchymal transition (164), activation of YAP/TAZ signaling (167), adeno-to-squamous cell carcinoma transition (87), and mucinous differentiation (170).
 Figure 4
Figure 4Resistance mechanisms to KRASG12C inhibitors and combination strategies. (A) Sequencing of circulating tumor DNA from patients who relapsed on adagrasib, sotorasib, divarasib, or LY3537982 treatment demonstrated that genetic alterations occurred at the level of RAS or in the upstream and downstream components of RAS signaling. RAS-level alterations included mutations and/or amplifications in KRAS and NRAS and mutations in NF1. Upstream signaling alterations included mutations, amplifications, and fusions in RTKs. Downstream signaling alterations included mutational activation of downstream ERK MAPK and PI3K effector signaling components, amplification of MYC, etc. No genetic mutations were found in 50% of patients who relapsed on KRASG12C treatment. (B) Most combination strategies with KRAS inhibitors are based on resistance mechanisms that have been identified in relapsed patients and in preclinical studies that include signal transduction and kinase inhibitors, among others (Tables 3 and 4). (C) Nongenetic mechanisms driving resistance to KRAS inhibitors may include transcriptional reprogramming, changes in cellular states (epithelial to mesenchymal [EMT], adeno-to-squamous carcinoma, or adenocarcinoma to mucinous differentiation), and/or changes in molecular subtypes. MET,mesenchymal to epithelial transition; RASi, RAS inhibitor.
The emerging complex resistance mechanisms suggest that combination strategies will be essential to improve the depth and duration of response to KRAS inhibitors. Guided in part by genetic alterations associated with relapsed tumors, and by preclinical CRISPR genetic screens or experimentally induced resistance assays, multiple combinations with KRAS inhibitors are currently under clinical evaluation (Figure 4B, Tables 3 and 4, and Supplemental Table 1). To date, the most promising combinations have involved inhibitors of upstream RTKs, particularly EGFR. The combination of anti-EGFR monoclonal antibodies with adagrasib and sotorasib led to the approval of these combinations for KRASG12C-mutant CRC (171, 172). Other combinations with inhibitors of additional components of the RAS signaling network (e.g., SOS1, ref. 173), immune checkpoint inhibitors (e.g., anti–PD-1 monoclonal antibodies, ref. 174), standard-of-care chemotherapy (e.g., gemcitabine/nab-paclitaxel, ref. 164), and co-occurring genetic alterations (MTAP-deletions with PRMT5 inhibitors, ref. 175) are currently under evaluation (Tables 3 and 4 and Supplemental Table 1).
-
Conclusions and future directions
After nearly four decades of effort, where many initially promising ideas failed to deliver clinically effective anti-KRAS therapies, the shattering of the myth that KRAS is undruggable has brought exciting new optimism that KRAS inhibitors will finally provide a significant therapeutic breakthrough in the treatment of PDAC. It is now conceivable that KRAS inhibitors may replace ineffective cytotoxic drugs as the standard of care. However, rather than marking the end of the road for anti-KRAS drug discovery, it is clearly early days in the process. The “best” class of KRAS inhibitors remains to be determined: mutation-selective, pan-KRAS or pan-RAS, allosteric small molecules versus degraders. The complex nature of mechanisms of resistance is arguably the most daunting challenge, highlighting the importance of continuing to develop other therapeutic strategies beyond direct KRAS inhibitors as well as to identify multiple effective combination therapies. Additional mutation-selective strategies, in particular for G12R and Q61X patients, may be needed. Biomarkers to identify patients who will respond to KRAS inhibitors and to monitor the efficacy of target inhibition will also be important.
The history of anti-KRAS drug discovery has been marked by misconceptions and an incomplete understanding of KRAS function, where concepts once considered dogma were later smashed and replaced. Although KRAS is one of the most intensely studied oncogenes, much remains to be understood about how it functions as a cancer driver. Despite these challenges, the discovery of KRAS inhibitors has ushered in a time of cautious optimism that the upward rate of pancreatic cancer deaths and the incremental steps in improvements to the 5-year survival rate of PDAC may soon be in our past.
-
Acknowledgments
ADC and CJD were supported by grants from the National Cancer Institute (NCI; R01CA42978, P50CA196510, P50CA257911, U01CA199235, P01CA203657, and R35CA232113), and CJD was supported by grants from the Pancreatic Cancer Action Network/American Association for Cancer Research (15-90-25-DER), the Pancreatic Cancer Action Network (22-WG-DERB), and the Department of Defense (W81XWH2110692). KDM was supported by NCI grant T32CA009156 and the American Cancer Society (PF-22-066-01-TBE).
Address correspondence to: Channing J. Der, University of North Carolina at Chapel Hill, Lineberger Comprehensive Cancer Center, Chapel Hill, North Carolina 27599, USA. Email: cjder@med.unc.edu.
-
Footnotes
Conflict of interest: ADC has consulted for Mirati Therapeutics Inc., a Bristol Myers Squibb company. CJD is a consultant/advisory board member for AskY Therapeutics; Cullgen; Deciphera Pharmaceuticals; Kestrel Therapeutics; Mirati Therapeutics Inc., a Bristol Myers Squibb company; Reactive Biosciences; Revolution Medicines; and SHY Therapeutics and has received research funding support from Deciphera Pharmaceuticals; Mirati Therapeutics Inc., a Bristol Myers Squibb company; Reactive Biosciences; Revolution Medicines; and SpringWorks Therapeutics.
Copyright: © 2025, Drizyte-Miller et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2025;135(16):e191939. https://doi.org/10.1172/JCI191939.
-
References
- Siegel RL, et al. Cancer statistics, 2025. CA Cancer J Clin. 2025;75(1):10–45.
- Rahib L, et al. Estimated projection of US cancer incidence and death to 2040. JAMA Netw Open. 2021;4(4):e214708.
- de Wilde RF, et al. Well-differentiated pancreatic neuroendocrine tumors: from genetics to therapy. Nat Rev Gastroenterol Hepatol. 2012;9(4):199–208.
- Hayashi A, et al. The pancreatic cancer genome revisited. Nat Rev Gastroenterol Hepatol. 2021;18(7):469–481.
- Sohal DPS, et al. Metastatic pancreatic cancer: ASCO guideline update. J Clin Oncol. 2020;38(27):3217–3230.
- Kleeff J, et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022.
- Conroy T, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364(19):1817–1825.
- Von Hoff DD, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691–1703.
- Der CJ, et al. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci U S A. 1982;79(11):3637–3640.
- Pulciani S, et al. Oncogenes in solid human tumours. Nature. 1982;300:539–542.
- Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–450.
- Collins MA, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122(2):639–653.
- Collins MA, et al. Metastatic pancreatic cancer is dependent on oncogenic Kras in mice. PLoS One. 2012;7(12):e49707.
- Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–670.
- Dang CV, et al. Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer. 2017;17(8):502–508.
- Cox AD, Der CJ. “Undruggable KRAS”: druggable after all. Genes Dev. 2025;39(1-2):132–62.
- Janne PA, et al. Adagrasib in non-small-cell lung cancer harboring a KRASG12C mutation. N Engl J Med. 2022;387(2):120–131.
- Skoulidis F, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N Engl J Med. 2021;384(25):2371–2381.
- Isermann T, et al. KRAS inhibitors: resistance drivers and combinatorial strategies. Trends Cancer. 2024;11(2):91–116.
- Singhi AD, et al. Early detection of pancreatic cancer: opportunities and challenges. Gastroenterology. 2019;156(7):2024–2040.
- Maitra A, et al. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol. 2003;16(9):902–912.
- Graham S, et al. From precursor to cancer: decoding the intrinsic and extrinsic pathways of pancreatic intraepithelial neoplasia progression. Carcinogenesis. 2024;45(11):801–816.
- Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32(2):185–203.
- Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806.
- Mullen KM, et al. The evolutionary forest of pancreatic cancer. Cancer Discov. 2025;15(2):329–345.
- Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501.
- Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52.
- Dentro SC, et al. Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell. 2021;184(8):2239–2254.
- Gerstung M, et al. Author Correction: The evolutionary history of 2,658 cancers. Nature. 2023;614(7948):E42.
- Notta F, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538(7625):378–382.
- Witkiewicz AK, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744.
- Flowers BM, et al. Cell of origin influences pancreatic cancer subtype. Cancer Discov. 2021;11(3):660–677.
- Varghese AM, et al. Clinicogenomic landscape of pancreatic adenocarcinoma identifies KRAS mutant dosage as prognostic of overall survival. Nat Med. 2025;31(2):466–477.
- Gopinathan A, et al. GEMMs as preclinical models for testing pancreatic cancer therapies. Dis Model Mech. 2015;8(10):1185–1200.
- Guerra C, Barbacid M. Genetically engineered mouse models of pancreatic adenocarcinoma. Mol Oncol. 2013;7(2):232–247.
- Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–483.
- Aguirre AJ, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–3126.
- Bardeesy N, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20(22):3130–3146.
- Basturk O, et al. A revised classification system and recommendations from the Baltimore Consensus Meeting for Neoplastic Precursor Lesions in the Pancreas. Am J Surg Pathol. 2015;39(12):1730–1741.
- Braxton AM, et al. 3D genomic mapping reveals multifocality of human pancreatic precancers. Nature. 2024;629(8012):679–687.
- Hosoda W, et al. Genetic analyses of isolated high-grade pancreatic intraepithelial neoplasia (HG-PanIN) reveal paucity of alterations in TP53 and SMAD4. J Pathol. 2017;242(1):16–23.
- Schutte M, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57(15):3126–3130.View this article via: PubMed Google Scholar
- Kanda M, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142(4):730–733.
- Matsuda Y, et al. The prevalence and clinicopathological characteristics of high-grade pancreatic intraepithelial neoplasia: autopsy study evaluating the entire pancreatic parenchyma. Pancreas. 2017;46(5):658–664.
- Brummelkamp TR, et al. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2(3):243–247.
- Singh A, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15(6):489–500.
- Prior IA, et al. The frequency of ras mutations in cancer. Cancer Res. 2020;80(14):2969–2974.
- Consortium APG. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7(8):818–831.
- Cox AD, et al. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13(11):828–851.
- Li S, et al. A model for RAS mutation patterns in cancers: finding the sweet spot. Nat Rev Cancer. 2018;18(12):767–777.
- Punekar SR, et al. The current state of the art and future trends in RAS-targeted cancer therapies. Nat Rev Clin Oncol. 2022;19(10):637–655.
- McIntyre CA, et al. Distinct clinical outcomes and biological features of specific KRAS mutants in human pancreatic cancer. Cancer Cell. 2024;42(9):1614–1629.
- Salem ME, et al. Landscape of KRASG12C, associated genomic alterations, and interrelation with immuno-oncology biomarkers in KRAS-mutated cancers. JCO Precis Oncol. 2022;6:e2100245.
- Haigis KM. KRAS alleles: the devil is in the detail. Trends Cancer. 2017;3(10):686–697.
- Cook JH, et al. The origins and genetic interactions of KRAS mutations are allele- and tissue-specific. Nat Commun. 2021;12(1):1808.
- Huynh MV, et al. Functional and biological heterogeneity of KRASQ61 mutations. Sci Signal. 2022;15(746):eabn2694.
- Johnson C, et al. Classification of KRAS-activating mutations and the implications for therapeutic intervention. Cancer Discov. 2022;12(4):913–923.
- Zafra MP, et al. An in vivo Kras allelic series reveals distinct phenotypes of common oncogenic variants. Cancer Discov. 2020;10(11):1654–1671.
- Hobbs GA, et al. Atypical KRASG12R mutant is impaired in PI3K signaling and macropinocytosis in pancreatic cancer. Cancer Discov. 2020;10(1):104–123.
- Commisso C, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497(7451):633–637.
- Fey SK, et al. KRAS loss of heterozygosity promotes MAPK-dependent pancreatic ductal adenocarcinoma initiation and induces therapeutic sensitivity to MEK inhibition. Cancer Res. 2025;85(2):251–262.
- Yan H, et al. Loss of the wild-type KRAS allele promotes pancreatic cancer progression through functional activation of YAP1. Oncogene. 2021;40(50):6759–6771.
- Zhang Z, et al. Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nat Genet. 2001;29(1):25–33.
- McGrath JP, et al. Structure and organization of the human Ki-ras proto-oncogene and a related processed pseudogene. Nature. 1983;304(5926):501–506.
- Bourne HR, et al. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348(6297):125–132.
- Bourne HR, et al. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349(6305):117–127.
- Wennerberg K, et al. The Ras superfamily at a glance. J Cell Sci. 2005;118(pt 5):843–846.
- Simanshu DK, et al. RAS proteins and their regulators in human disease. Cell. 2017;170(1):17–33.
- Bos JL, et al. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129(5):865–877.
- Vigil D, et al. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10(12):842–857.
- Hunter JC, et al. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol Cancer Res. 2015;13(9):1325–1335.
- Catozzi S, et al. Predicted ‘wiring landscape’ of Ras-effector interactions in 29 human tissues. NPJ Syst Biol Appl. 2021;7(1):10.
- Kiel C, et al. The ins and outs of RAS effector complexes. Biomolecules. 2021;11(2):236.
- Drosten M, Barbacid M. Targeting the MAPK pathway in KRAS-driven tumors. Cancer Cell. 2020;37(4):543–550.
- Lavoie H, et al. ERK signalling: a master regulator of cell behaviour, life and fate. Nat Rev Mol Cell Biol. 2020;21(10):607–632.
- Klomp JE, et al. Determining the ERK-regulated phosphoproteome driving KRAS-mutant cancer. Science. 2024;384(6700):eadk0850.
- Klomp JA, et al. Defining the KRAS- and ERK-dependent transcriptome in KRAS-mutant cancers. Science. 2024;384(6700):eadk0775.
- Wolpaw AJ, Dang CV. MYC-induced metabolic stress and tumorigenesis. Biochim Biophys Acta Rev Cancer. 2018;1870(1):43–50.
- Ozkan-Dagliyan I, et al. Low-dose vertical inhibition of the RAF-MEK-ERK cascade causes apoptotic death of KRAS mutant cancers. Cell Rep. 2020;31(11):107764.
- Vaseva AV, et al. KRAS suppression-induced degradation of MYC is antagonized by a MEK5-ERK5 compensatory mechanism. Cancer Cell. 2018;34(5):807–822.
- Sodir NM, et al. MYC instructs and maintains pancreatic adenocarcinoma phenotype. Cancer Discov. 2020;10(4):588–607.
- Vasan N, Cantley LC. At a crossroads: how to translate the roles of PI3K in oncogenic and metabolic signalling into improvements in cancer therapy. Nat Rev Clin Oncol. 2022;19(7):471–485.
- Murthy D, et al. Phosphoinositide 3-kinase signaling pathway in pancreatic ductal adenocarcinoma progression, pathogenesis, and therapeutics. Front Physiol. 2018;9:335.
- Castellano E, et al. Requirement for interaction of PI3-kinase p110α with RAS in lung tumor maintenance. Cancer Cell. 2013;24(5):617–630.
- Gupta S, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129(5):957–968.
- Collisson EA, et al. A central role for RAF→MEK→ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2012;2(8):685–693.
- Awad MM, et al. Acquired resistance to KRASG12C inhibition in cancer. N Engl J Med. 2021;384(25):2382–2393.
- Zhao Y, et al. Diverse alterations associated with resistance to KRAS(G12C) inhibition. Nature. 2021;599(7886):679–683.
- Macdonald JS, et al. A phase II study of farnesyl transferase inhibitor R115777 in pancreatic cancer: a Southwest oncology group (SWOG 9924) study. Invest New Drugs. 2005;23(5):485–487.
- Van Cutsem E, et al. Phase III trial of gemcitabine plus tipifarnib compared with gemcitabine plus placebo in advanced pancreatic cancer. J Clin Oncol. 2004;22(8):1430–1438.
- Cohen SJ, et al. Phase II and pharmacodynamic study of the farnesyltransferase inhibitor R115777 as initial therapy in patients with metastatic pancreatic adenocarcinoma. J Clin Oncol. 2003;21(7):1301–1306.
- James G, et al. Resistance of K-RasBV12 proteins to farnesyltransferase inhibitors in Rat1 cells. Proc Natl Acad Sci U S A. 1996;93(9):4454–4458.
- Rowell CA, et al. Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. J Biol Chem. 1997;272(22):14093–14097.
- Whyte DB, et al. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem. 1997;272(22):14459–14464.
- Kohl NE, et al. Selective inhibition of ras-dependent transformation by a farnesyltransferase inhibitor. Science. 1993;260(5116):1934–1937.
- Kohl NE, et al. Inhibition of farnesyltransferase induces regression of mammary and salivary carcinomas in ras transgenic mice. Nat Med. 1995;1(8):792–797.
- Hussain S, et al. Targeting oncogenic kinases: Insights on FDA approved tyrosine kinase inhibitors. Eur J Pharmacol. 2024;970:176484.
- Roskoski R, JrProperties of FDA-approved small molecule protein kinase inhibitors: A 2024 update. Pharmacol Res. 2024;200:107059.
- Klomp JE, et al. The ERK mitogen-activated protein kinase signaling network: the final frontier in RAS signal transduction. Biochem Soc Trans. 2021;49(1):253–267.
- Ryan MB, et al. Targeting RAS-mutant cancers: is ERK the key? Trends Cancer. 2015;1(3):183–198.
- Gouda MA, Subbiah V. Precision oncology for BRAF-mutant cancers with BRAF and MEK inhibitors: from melanoma to tissue-agnostic therapy. ESMO Open. 2023;8(2):100788.
- Moertel CL, et al. ReNeu: a pivotal, phase IIb trial of mirdametinib in adults and children with symptomatic neurofibromatosis type 1-associated plexiform neurofibroma. J Clin Oncol. 2025;43(6):716–729.
- Grierson PM, et al. Phase Ib study of ulixertinib plus gemcitabine and nab-paclitaxel in patients with metastatic pancreatic adenocarcinoma. Oncologist. 2023;28(2):115–123.
- Mortazavi M, et al. Prospects of targeting PI3K/AKT/mTOR pathway in pancreatic cancer. Crit Rev Oncol Hematol. 2022;176:103749.
- Bryant KL, et al. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39(2):91–100.
- DeLiberty JM, et al. Unraveling and targeting RAS-driven metabolic signaling for therapeutic gain. Adv Cancer Res. 2022;153:267–304.
- Zeh HJ, et al. A Randomized phase II preoperative study of autophagy inhibition with high-dose hydroxychloroquine and gemcitabine/nab-paclitaxel in pancreatic cancer patients. Clin Cancer Res. 2020;26(13):3126–3134.
- Bryant KL, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med. 2019;25(4):628–640.
- Kinsey CG, et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 2019;25(4):620–627.
- Philip PA, et al. Devimistat (CPI-613) with modified fluorouarcil, oxaliplatin, irinotecan, and leucovorin (FFX) versus FFX for patients with metastatic adenocarcinoma of the pancreas: The Phase III AVENGER 500 Study. J Clin Oncol. 2024;42(31):3692–3701.
- Biancur DE, et al. Compensatory metabolic networks in pancreatic cancers upon perturbation of glutamine metabolism. Nat Commun. 2017;8:15965.
- Cheng C, et al. Targeting PIKfyve-driven lipid metabolism in pancreatic cancer. Nature. 2025;642(8068):776–784.
- DeLiberty JM, et al. Concurrent inhibition of the RAS-MAPK pathway and PIKfyve is a therapeutic strategy for pancreatic cancer. Cancer Res. 2025;85(8):1479–1495.
- Feuerstein J, et al. Characterisation of the metal-ion-GDP complex at the active sites of transforming and nontransforming p21 proteins by observation of the 17O-Mn superhyperfine coupling and by kinetic methods. Eur J Biochem. 1987;162(1):49–55.
- Pai EF, et al. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989;341(6239):209–214.
- Ostrem JM, et al. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503(7477):548–551.
- Ostrem JML, et al. Direct RAS inhibitors turn 10. Nat Chem Biol. 2024;20(10):1238–1241.
- Lanman BA, et al. Discovery of a covalent inhibitor of KRASG12C (AMG 510) for the treatment of solid tumors. J Med Chem. 2020;63(1):52–65.
- Hallin J, et al. The KRASG12C Inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020;10(1):54–71.
- Hong DS, et al. KRASG12C inhibition with sotorasib in advanced solid tumors. N Engl J Med. 2020;383(13):1207–1217.
- de Langen AJ, et al. Sotorasib versus docetaxel for previously treated non-small-cell lung cancer with KRASG12C mutation: a randomised, open-label, phase 3 trial. Lancet. 2023;401(10378):733–746.
- Mok TSK, et al. KRYSTAL-12: Phase 3 study of adagrasib versus docetaxel in patients with previously treated advanced/metastatic non-small cell lung cancer (NSCLC) harboring a KRASG12C mutation. J Clin Oncol. 2024;42(17_suppl):LBA8509. View this article via: CrossRef Google Scholar
- Strickler JH, et al. Sotorasib in KRAS p.G12C-mutated advanced pancreatic cancer. N Engl J Med. 2023;388(1):33–43.
- Bekaii-Saab TS, et al. Adagrasib in advanced solid tumors harboring a KRASG12C mutation. J Clin Oncol. 2023;41(25):4097–4106.
- Sacher A, et al. Single-agent divarasib (GDC-6036) in solid tumors with a KRAS G12C mutation. N Engl J Med. 2023;389(8):710–721.
- Maciag AE, et al. Discovery of BBO-8520, a first-in-class direct and covalent dual inhibitor of GTP-Bound (ON) and GDP-bound (OFF) KRASG12C. Cancer Discov. 2024;15(3):578–594.
- Schulze CJ, et al. Chemical remodeling of a cellular chaperone to target the active state of mutant KRAS. Science. 2023;381(6659):794–799.
- Zhang Z, et al. A covalent inhibitor of K-Ras(G12C) induces MHC class I presentation of haptenated peptide neoepitopes targetable by immunotherapy. Cancer Cell. 2022;40(9):1060–1069.
- Hattori T, et al. Creating MHC-restricted neoantigens with covalent inhibitors that can be targeted by immune therapy. Cancer Discov. 2023;13(1):132–145.
- Hallin J, et al. Anti-tumor efficacy of a potent and selective non-covalent KRASG12D inhibitor. Nat Med. 2022;28(10):2171–2182.
- Wang X, et al. Identification of MRTX1133, a noncovalent, potent, and selective KRASG12D inhibitor. J Med Chem. 2022;65(4):3123–3133.
- Kemp SB, et al. Efficacy of a small-molecule inhibitor of KrasG12D in immunocompetent models of pancreatic cancer. Cancer Discov. 2023;13(2):298–311.
- Jiang L, et al. Abstract 526: RMC-9805, a first-in-class, mutant-selective, covalent and oral KRASG12D(ON) inhibitor that induces apoptosis and drives tumor regression in preclinical models of KRASG12D cancers. Cancer Res. 2023;83(7_suppl):526. View this article via: CrossRef Google Scholar
- Gong X, et al. Abstract 3316: LY3962673, an oral, highly potent, mutant-selective, and non-covalent KRAS G12D inhibitor demonstrates robust anti-tumor activity in KRAS G12D models. Cancer Res. 2024;84(6_suppl):3316. View this article via: CrossRef Google Scholar
- Vo ED, et al. Abstract LB321: Discovery and characterization of QTX3046, a potent, selective, and orally bioavailable non-covalent KRASG12D inhibitor. Cancer Res. 2023;83(8_suppl):LB321. View this article via: CrossRef Google Scholar
- Yan F, et al. Abstract 3318: GFH375 (VS-7375): An oral, selective KRAS G12D (ON/OFF) inhibitor with potent anti-tumor efficacy. Cancer Res. 2024;84(6_suppl):3318. View this article via: CrossRef Google Scholar
- Zhou C, et al. Anti-tumor efficacy of HRS-4642 and its potential combination with proteasome inhibition in KRAS G12D-mutant cancer. Cancer Cell. 2024;42(7):1286–1300.
- Shang E, et al. Abstract 3315: Preclinical studies of TSN1611, a potent, selective, and orally bioavailable KRASG12D inhibitor. Cancer Res. 2024;84(6_suppl):3315. View this article via: CrossRef Google Scholar
- Farren MR, et al. Abstract 5900: INCB161734: A novel, potent, and orally bioavailable KRAS G12D selective inhibitor demonstrates antitumor activity in KRAS G12D mutant tumors. Cancer Res. 2024;84(6_suppl):5900. View this article via: CrossRef Google Scholar
- Kim D, et al. Pan-KRAS inhibitor disables oncogenic signalling and tumour growth. Nature. 2023;619(7968):160–166.
- Tedeschi A, et al. Pan-KRAS inhibitors BI-2493 and BI-2865 display potent antitumor activity in tumors with KRAS wild-type allele amplification. Mol Cancer Ther. 2024;24(4):550–562.
- Zhang YW, et al. Abstract LB320: Discovery and characterization of QTX3034, a potent, selective, and orally bioavailable allosteric KRAS inhibitor. Cancer Res. 2023;83(8_suppl):LB320. View this article via: CrossRef Google Scholar
- Damnernsawad A, et al. Kras is required for adult hematopoiesis. Stem Cells. 2016;34(7):1859–1871.
- Fuentes-Mateos R, et al. Combined HRAS and NRAS ablation induces a RASopathy phenotype in mice. Cell Commun Signal. 2024;22(1):332.
- Fuentes-Mateos R, et al. Concomitant deletion of HRAS and NRAS leads to pulmonary immaturity, respiratory failure and neonatal death in mice. Cell Death Dis. 2019;10(11):838.
- Johnson L, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11(19):2468–2481.
- Koera K, et al. K-ras is essential for the development of the mouse embryo. Oncogene. 1997;15(10):1151–1159.
- Holderfield M, et al. Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy. Nature. 2024;629(8013):919–926.
- Jiang J, et al. Translational and therapeutic evaluation of RAS-GTP Inhibition by RMC-6236 in RAS-driven cancers. Cancer Discov. 2024;14(6):994–1017.
- Wasko UN, et al. Tumour-selective activity of RAS-GTP inhibition in pancreatic cancer. Nature. 2024;629(8013):927–936.
- Cuevas-Navarro A, et al. Pharmacological restoration of GTP hydrolysis by mutant RAS. Nature. 2025;637(8044):224–229.
- Ryan MB, et al. Vertical pathway inhibition overcomes adaptive feedback resistance to KRASG12C inhibition. Clin Cancer Res. 2020;26(7):1633–1643.
- Garrido-Laguna I, et al. Safety, efficacy, and on-treatment circulating tumor DNA (ctDNA) changes from a phase 1 study of RMC-6236, a RAS(ON) multi-selective, tri-complex inhibitor, in patients with RAS mutant pancreatic ductal adenocarcinoma (PDAC). J Clin Oncol. 2025;43(4_suppl):722. View this article via: CrossRef Google Scholar
- Golan T, et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget. 2015;6(27):24560–24570.
- Nagashima T, et al. Abstract 5735: Novel KRAS G12D degrader ASP3082 demonstrates in vivo, dose-dependent KRAS degradation, KRAS pathway inhibition, and antitumor efficacy in multiple KRAS G12D-mutated cancer models. Cancer Res. 2023;83(7_suppl):5735. View this article via: CrossRef Google Scholar
- Park W, et al. 608O Preliminary safety and clinical activity of ASP3082, a first-in-class, KRAS G12D selective protein degrader in adults with advanced pancreatic (PC), colorectal (CRC), and non-small cell lung cancer (NSCLC). Ann Oncol. 2024;35(suppl_2):S486–S487.View this article via: CrossRef Google Scholar
- Popow J, et al. Targeting cancer with small-molecule pan-KRAS degraders. Science. 2024;385(6715):1338–1347.
- Palmer DH, et al. TG01/GM-CSF and adjuvant gemcitabine in patients with resected RAS-mutant adenocarcinoma of the pancreas (CT TG01-01): a single-arm, phase 1/2 trial. Br J Cancer. 2020;122(7):971–977.
- Nagasaka M, et al. KRAS Inhibitors- yes but what next? Direct targeting of KRAS- vaccines, adoptive T cell therapy and beyond. Cancer Treat Rev. 2021;101:102309.
- Pant S, et al. Lymph-node-targeted, mKRAS-specific amphiphile vaccine in pancreatic and colorectal cancer: the phase 1 AMPLIFY-201 trial. Nat Med. 2024;30(2):531–542.
- Belnoue E, et al. Targeting self and neo-epitopes with a modular self-adjuvanting cancer vaccine. JCI Insight. 2019;5(11):e127305.
- Bear AS, et al. Natural TCRs targeting KRASG12V display fine specificity and sensitivity to human solid tumors. J Clin Invest. 2024;134(21):e175790.
- Rojas LA, et al. Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature. 2023;618(7963):144–150.
- Dilly J, et al. Mechanisms of resistance to oncogenic KRAS inhibition in pancreatic cancer. Cancer Discov. 2024;14(11):2135–2161.
- Tanaka N, et al. Clinical acquired resistance to KRASG12C inhibition through a novel KRAS Switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov. 2021;11(8):1913–1922.
- Yaeger R, et al. Molecular characterization of acquired resistance to KRASG12C-EGFR inhibition in colorectal cancer. Cancer Discov. 2023;13(1):41–55.
- Tsai YS, et al. Rapid idiosyncratic mechanisms of clinical resistance to KRAS G12C inhibition. J Clin Invest. 2022;132(4):e155523.
- Desai J, et al. Divarasib plus cetuximab in KRAS G12C-positive colorectal cancer: a phase 1b trial. Nat Med. 2024;30(1):271–278.
- Riedl JM, et al. Genomic landscape of clinically acquired resistance alterations in patients treated with KRASG12C inhibitors. Ann Oncol. 2025;36(6):682–692.
- Araujo HA, et al. Mechanisms of response and tolerance to active RAS inhibition in KRAS-mutant non-small cell lung cancer. Cancer Discov. 2024;14(11):2183–2208.
- Fakih MG, et al. Sotorasib plus panitumumab in refractory colorectal cancer with mutated KRAS G12C. N Engl J Med. 2023;389(23):2125–2139.
- Yaeger R, et al. Efficacy and safety of adagrasib plus cetuximab in patients with KRASG12C-mutated metastatic colorectal cancer. Cancer Discov. 2024;14(6):982–993.
- Thatikonda V, et al. Co-targeting SOS1 enhances the antitumor effects of KRASG12C inhibitors by addressing intrinsic and acquired resistance. Nat Cancer. 2024;5(9):1352–1370.
- Briere DM, et al. The KRASG12C inhibitor MRTX849 reconditions the tumor immune microenvironment and sensitizes tumors to checkpoint inhibitor therapy. Mol Cancer Ther. 2021;20(6):975–985.
- Belmontes B, et al. AMG 193, a clinical stage MTA-cooperative PRMT5 inhibitor, drives antitumor activity preclinically and in patients with MTAP-deleted cancers. Cancer Discov. 2024;15(1):139–161.
- Connor AA, Gallinger S. Pancreatic cancer evolution and heterogeneity: integrating omics and clinical data. Nat Rev Cancer. 2022;22(3):131–142.
- Almoguera C, et al. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53(4):549–554.
- Blasco MT, et al. Complete regression of advanced pancreatic ductal adenocarcinomas upon combined inhibition of EGFR and C-RAF. Cancer Cell. 2019;35(4):573–587.
- Bodoky G, et al. A phase II open-label randomized study to assess the efficacy and safety of selumetinib (AZD6244 [ARRY-142886]) versus capecitabine in patients with advanced or metastatic pancreatic cancer who have failed first-line gemcitabine therapy. Invest New Drugs. 2012;30(3):1216–1223.
- Infante JR, et al. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur J Cancer. 2014;50(12):2072–2081.
- Chung V, et al. Effect of selumetinib and MK-2206 vs oxaliplatin and fluorouracil in patients with metastatic pancreatic cancer after prior therapy: SWOG S1115 Study Randomized Clinical Trial. JAMA Oncol. 2017;3(4):516–522.
- Van Cutsem E, et al. Phase I/II trial of pimasertib plus gemcitabine in patients with metastatic pancreatic cancer. Int J Cancer. 2018;143(8):2053–2064.
- Kenney C, et al. Phase II study of selumetinib, an orally active inhibitor of MEK1 and MEK2 kinases, in KRASG12R-mutant pancreatic ductal adenocarcinoma. Invest New Drugs. 2021;39(3):821–828.
- Arbour KC, et al. 652O Preliminary clinical activity of RMC-6236, a first-in-class, RAS-selective, tri-complex RAS-MULTI(ON) inhibitor in patients with KRAS mutant pancreatic ductal adenocarcinoma (PDAC) and non-small cell lung cancer (NSCLC). Ann Oncol. 2023;34(2_suppl):S458. View this article via: CrossRef Google Scholar
- Wolpin B, et al. 514LBA (PB-514) LBA Posters: Updated safety and efficacy from a Phase 1 study of RMC-6236, a RAS (ON) multi-selective, tri-complex inhibitor, in patients with RAS mutant pancreatic ductal adenocarcinoma (PDAC). Eur J Cancer. 2024;211:114992. View this article via: CrossRef Google Scholar
- Spira AI, et al. Preliminary safety, antitumor activity, and circulating tumor DNA (ctDNA) changes with RMC-9805, an oral, RAS(ON) G12D-selective tri-complex inhibitor in patients with KRAS G12D pancreatic ductal adenocarcinoma (PDAC) from a phase 1 study in advanced solid tumors. J Clin Oncol. 2025;43(4_suppl):724. View this article via: CrossRef Google Scholar
- Gupta A, et al. Clinical outcomes of liposomal irinotecan in advanced pancreatic adenocarcinoma patients previously treated with conventional irinotecan-based chemotherapy: a real-world study. Front Oncol. 2023;13:1250136.
- Wang-Gillam A, et al. Nanoliposomal irinotecan with fluorouracil and folinic acid in metastatic pancreatic cancer after previous gemcitabine-based therapy (NAPOLI-1): a global, randomised, open-label, phase 3 trial. Lancet. 2016;387(10018):545–557.
- Chiorean EG, et al. Randomized phase II study of PARP inhibitor ABT-888 (Veliparib) with modified FOLFIRI versus FOLFIRI as second-line treatment of metastatic pancreatic cancer: SWOG S1513. Clin Cancer Res. 2021;27(23):6314–6322.
- Hecht JR, et al. Randomized Phase III Study of FOLFOX alone or with pegilodecakin as second-line therapy in patients with metastatic pancreatic cancer that progressed after gemcitabine (SEQUOIA). J Clin Oncol. 2021;39(10):1108–1118.
- Huffman BM, et al. Effect of a MUC5AC antibody (NPC-1C) administered with second-line gemcitabine and nab-paclitaxel on the survival of patients with advanced pancreatic ductal adenocarcinoma: a randomized clinical trial. JAMA Netw Open. 2023;6(1):e2249720.
- Hammel P, et al. Trybeca-1: A randomized, phase 3 study of eryaspase in combination with chemotherapy versus chemotherapy alone as second-line treatment in patients with advanced pancreatic adenocarcinoma (NCT03665441). J Clin Oncol. 2022;40(4_suppl):518. View this article via: CrossRef Google Scholar
- De La Fouchardiere C, et al. Gemcitabine and paclitaxel versus gemcitabine alone after 5-fluorouracil, oxaliplatin, and irinotecan in metastatic pancreatic adenocarcinoma: a randomized phase III PRODIGE 65-UCGI 36-GEMPAX UNICANCER Study. J Clin Oncol. 2024;42(9):1055–1066.
- Schram AM, et al. Efficacy of zenocutuzumab in NRG1 fusion-positive cancer. N Engl J Med. 2025;392(6):566–576.
- Moore MJ, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25(15):1960–1966.
- Marabelle A, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 Study. J Clin Oncol. 2020;38(1):1–10.
- O’Reilly EM, et al. Randomized, multicenter, phase II trial of gemcitabine and cisplatin with or without veliparib in patients with pancreas adenocarcinoma and a germline BRCA/PALB2 mutation. J Clin Oncol. 2020;38(13):1378–1388.
- Wainberg ZA, et al. NALIRIFOX versus nab-paclitaxel and gemcitabine in treatment-naive patients with metastatic pancreatic ductal adenocarcinoma (NAPOLI 3): a randomised, open-label, phase 3 trial. Lancet. 2023;402(10409):1272–1281.
-
Version history
- Version 1 (August 15, 2025): Electronic publication
Article tools
-
View PDF

- Download citation information
- Send a comment
- Terms of use
- Standard abbreviations
- Need help? Email the journal
Review Series
Pancreatic Cancer
-
Pancreatic ductal adenocarcinoma: the Everest of cancer biology
Minh T. Than et al. -
Early neoplastic lesions of the pancreas: initiation, progression, and opportunities for precancer interception
Brian A. Pedro et al. -
KRAS: the Achilles’ heel of pancreas cancer biology
Kristina Drizyte-Miller et al. -
What’s on the menu?: metabolic constraints in the pancreatic tumor microenvironment
Colin Sheehan et al. -
Challenges of early detection of pancreatic cancer
Michael J. Shen et al. -
Metastatic heterogeneity in pancreatic cancer: mechanisms and opportunities for targeted intervention
Ravikanth Maddipati -
Evolving concepts in adjuvant/neoadjuvant therapy for resectable pancreas cancer
John M. Bryant et al. -
Experimental models of pancreas cancer: what has been the impact for precision medicine?
Vasiliki Pantazopoulou et al.
Metrics
Go to
- Top
- Abstract
- Introduction
- KRAS — the driver of pancreatic cancer
- KRAS mutations
- KRAS signaling in PDAC
- Early approaches for anti-KRAS therapies in PDAC
- Development of direct KRAS inhibitors
- Other KRAS therapeutic strategies
- Resistance to KRAS inhibitors and promising combination strategies
- Conclusions and future directions
- Supplemental material
- Acknowledgments
- Footnotes
- References
- Version history



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)








